dplyr
# install.packages("dplyr")
library(tidyverse)This document is based on package version of dplyr:
packageVersion("dplyr")
#> [1] '1.0.6'1 Why dplyr
When working with data you must:
Figure out what you want to do.
Describe those tasks in the form of a computer program.
Execute the program.
The dplyr package makes these steps fast and easy:
By constraining your options, it helps you think about your data manipulation challenges.
It provides simple “verbs”, functions that correspond to the most common data manipulation tasks, to help you translate your thoughts into code.
It uses efficient backends, so you spend less time waiting for the computer.
dplyr codes usually looks like:
starwars %>%
select(name, height, species) %>%
filter(species %in% c('Human', 'Droid')) %>%
mutate(mean_height = mean(height, na.rm = T)) %>%
group_by(species) %>%
mutate(mean_height_by_species = mean(height, na.rm = T)) %>%
ungroup()Here we use several dplyr “verbs” (select() filter() mutate()) to operate the “data” starwars step by step, and string them together into a pipeline by %>%. We also finish one “by-group” work by wrapping mutate() with group_by() and ungroup(). This code block looks like working left-to-right, top-to-bottom and you can easily find what is done to the data.
So how dplyr do this?
The code dplyr verbs input and output data frames.
dplyr relies heavily on “non-standard evaluation” so that you don’t need to use
$to refer to columns in the “current” data frame.dplyr solutions tend to use a variety of single purpose verbs.
Multiple dplyr verbs are often strung together into a pipeline by
%>%.All dplyr verbs handle “grouped” data frames so that the code to perform a computation per-group looks very similar to code that works on a whole data frame.
2 Tibble
A tibble, or tbl_df, is a modern reimagining of the data.frame, keeping what time has proven to be effective, and throwing out what is not. Tibbles are data.frames that are lazy and surly: they do less (i.e. they don’t change variable names or types, and don’t do partial matching) and complain more (e.g. when a variable does not exist). This forces you to confront problems earlier, typically leading to cleaner, more expressive code. Tibbles also have an enhanced print() method which makes them easier to use with large datasets containing complex objects.
mtcars %>% class()
#> [1] "data.frame"
mtcars %>% as_tibble() %>% class()
#> [1] "tbl_df" "tbl" "data.frame"You can refer to tibbles chapter in R for data science or vignette("tibble") to find the difference between tibble and data.frame.
Tibble is the central data structure for the set of packages known as the tidyverse, including dplyr, ggplot2, tidyr, and readr.
2.1 Creating tibbles
Create a tibble from an existing object with as_tibble(). This will work for reasonable inputs that are already data.frames, lists, matrices, or tables.
as_tibble(iris)
#> # A tibble: 150 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> # … with 146 more rowsYou can also create a new tibble from column vectors with tibble(). tibble() will automatically recycle inputs of length 1, and allows you to refer to variables that you just created, as shown below.
tibble(x = 1:5, y = 1, z = x ^ 2 + y)
#> # A tibble: 5 x 3
#> x y z
#> <int> <dbl> <dbl>
#> 1 1 1 2
#> 2 2 1 5
#> 3 3 1 10
#> 4 4 1 17
#> # … with 1 more rowYou can define a tibble row-by-row with tribble(), short for transposed tibble. tribble() is customised for data entry in code: column headings are defined by formulas (i.e. they start with ~), and entries are separated by commas. This makes it possible to lay out small amounts of data in easy to read form.
tribble(
~x, ~y, ~z,
"a", 2, 3.6,
"b", 1, 8.5
)
#> # A tibble: 2 x 3
#> x y z
#> <chr> <dbl> <dbl>
#> 1 a 2 3.6
#> 2 b 1 8.52.2 Display
Tibbles have a refined print method that shows only the first 10 rows (we set it to 4 in this document), and all the columns that fit on screen (by default). This makes it much easier to work with large data. In addition to its name, each column reports its type, a nice feature borrowed from str():
starwars
#> # A tibble: 87 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>You can also control the default print behaviour by setting options:
options(tibble.print_max = n, tibble.print_min = m): if more than n rows, print only m rows. Useoptions(tibble.print_min = Inf)to always show all rows.Use
options(tibble.width = Inf)to always print all columns, regardless of the width of the screen.
You can see a complete list of options by looking at the package help with package?tibble.
A final option is to use RStudio’s built-in data viewer to get a scrollable view of the complete dataset. This is also often useful at the end of a long chain of manipulations.
starwars %>% view()2.3 Column data types
Refer to vignette("types"), you can find all kinds of column data type in tibble, common are <lgl> for logical, <int> for integer, <dbl> for double, <chr> for character, <date> for Date and so on.
tibble(x1 = T, x2 = 1L, x3 = 1.5, x4 = 'a', x5 = as.Date('2021-03-10'))
#> # A tibble: 1 x 5
#> x1 x2 x3 x4 x5
#> <lgl> <int> <dbl> <chr> <date>
#> 1 TRUE 1 1.5 a 2021-03-10And sometimes you would find <list> for list (list-columns).
tibble(x = list(1, 2:3))
#> # A tibble: 2 x 1
#> x
#> <list>
#> 1 <dbl [1]>
#> 2 <int [2]>
mtcars %>% nest_by(cyl)
#> # A tibble: 3 x 2
#> # Rowwise: cyl
#> cyl data
#> <dbl> <list<tibble[,10]>>
#> 1 4 [11 × 10]
#> 2 6 [7 × 10]
#> 3 8 [14 × 10]list-columns might be confusing but sometimes it would be useful in modeling while the input are multi tibbles, you can see more details in vignette("rowwise") and Many models chapter in R for data science. Later we will also meet list-columns with “nest” data while study the package tidyr.
2.4 Column names
The name repair described below is exposed to users via the .name_repair argument of tibble::tibble(), tibble::as_tibble(), readxl::read_excel(), and, eventually other packages in the tidyverse.
Tidyverse deals with a few levels of name repair that are practically useful:
minimal: Thenamesattribute is notNULL. The name of an unnamed element is""(the empty string) and neverNA.unique: No element ofnamesappears more than once. A couple specific names are also forbidden in unique names, such as""(the empty string).- All columns can be accessed by name via
df[["name"]]and, more generally, by quoting with backticks:df$`name`,subset(df, select = `name`), anddplyr::select(df, `name`).
- All columns can be accessed by name via
check_unique: It doesn’t perform any name repair. Instead, an error is raised if the names don’t suit the “unique” criteria.universal: Thenamesare unique and syntactic.- Names work everywhere, without quoting:
df$nameandlm(name1 ~ name2, data = df)anddplyr::select(df, name)all work.
- Names work everywhere, without quoting:
c('a', '', NA) %>% vctrs::vec_as_names(repair = 'minimal')
#> [1] "a" "" ""
c('a', 'b', 'c', 'a') %>% vctrs::vec_as_names(repair = 'unique')
#> New names:
#> * a -> a...1
#> * a -> a...4
#> [1] "a...1" "b" "c" "a...4"
c('a', 'b', 'c', 'a') %>% vctrs::vec_as_names(repair = 'check_unique')
#> Error: Names must be unique.
#> x These names are duplicated:
#> * "a" at locations 1 and 4.
c('(a)', 'b c', '') %>% vctrs::vec_as_names(repair = 'universal')
#> New names:
#> * `(a)` -> .a.
#> * `b c` -> b.c
#> * `` -> ...3
#> [1] ".a." "b.c" "...3"You can see more details in ?vctrs::vec_as_names and Names attribute chapter in Tidyverse design guide.
3 Single table verbs
3.1 Arrange rows by column values: arrange()
arrange() orders the rows of a data frame by the values of selected columns.
starwars %>% arrange(height, mass)
#> # A tibble: 87 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Yoda 66 17 white green brown 896 male mascu…
#> 2 Ratts Ty… 79 15 none grey, blue unknown NA male mascu…
#> 3 Wicket S… 88 20 brown brown brown 8 male mascu…
#> 4 Dud Bolt 94 45 none blue, grey yellow NA male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>% arrange(desc(height))
#> # A tibble: 87 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Yarael … 264 NA none white yellow NA male mascul…
#> 2 Tarfful 234 136 brown brown blue NA male mascul…
#> 3 Lama Su 229 88 none grey black NA male mascul…
#> 4 Chewbac… 228 112 brown unknown blue 200 male mascul…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>3.2 Subset distinct/unique rows: distinct()
distinct() select only unique/distinct rows from a data frame.
df <- tibble(
x = sample(10, 100, rep = TRUE),
y = sample(10, 100, rep = TRUE)
)
df
#> # A tibble: 100 x 2
#> x y
#> <int> <int>
#> 1 6 8
#> 2 8 2
#> 3 5 8
#> 4 3 6
#> # … with 96 more rows
df %>% nrow()
#> [1] 100
df %>% distinct()
#> # A tibble: 64 x 2
#> x y
#> <int> <int>
#> 1 6 8
#> 2 8 2
#> 3 5 8
#> 4 3 6
#> # … with 60 more rows
df %>% distinct() %>% nrow()
#> [1] 64
df %>% distinct(x)
#> # A tibble: 10 x 1
#> x
#> <int>
#> 1 6
#> 2 8
#> 3 5
#> 4 3
#> # … with 6 more rows
df %>% distinct(y)
#> # A tibble: 10 x 1
#> y
#> <int>
#> 1 8
#> 2 2
#> 3 6
#> 4 1
#> # … with 6 more rows
df %>% distinct(x, .keep_all = T)
#> # A tibble: 10 x 2
#> x y
#> <int> <int>
#> 1 6 8
#> 2 8 2
#> 3 5 8
#> 4 3 6
#> # … with 6 more rows
df %>% distinct(y, .keep_all = T)
#> # A tibble: 10 x 2
#> x y
#> <int> <int>
#> 1 6 8
#> 2 8 2
#> 3 3 6
#> 4 2 1
#> # … with 6 more rows3.3 Subset rows using column values: filter()
The filter() function is used to subset a data frame, retaining all rows that satisfy your conditions. To be retained, the row must produce a value of TRUE for all conditions. Note that when a condition evaluates to NA the row will be dropped.
starwars %>%
filter(hair_color == "none" & eye_color == "black")
#> # A tibble: 9 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Nien N… 160 68 none grey black NA male mascul…
#> 2 Gasgano 122 NA none white, blue black NA male mascul…
#> 3 Kit Fi… 196 87 none green black NA male mascul…
#> 4 Plo Ko… 188 80 none orange black 22 male mascul…
#> # … with 5 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>%
filter(hair_color == "none" | eye_color == "black")
#> # A tibble: 38 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Darth V… 202 136 none white yellow 41.9 male mascul…
#> 2 Greedo 173 74 <NA> green black 44 male mascul…
#> 3 IG-88 200 140 none metal red 15 none mascul…
#> 4 Bossk 190 113 none green red 53 male mascul…
#> # … with 34 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>3.4 Subset columns using their names and types: select()
select() select variables in a data frame
starwars %>% select(hair_color, skin_color, eye_color)
#> # A tibble: 87 x 3
#> hair_color skin_color eye_color
#> <chr> <chr> <chr>
#> 1 blond fair blue
#> 2 <NA> gold yellow
#> 3 <NA> white, blue red
#> 4 none white yellow
#> # … with 83 more rows3.5 Create, modify, and delete columns: mutate() transmute()
mutate() adds new variables and preserves existing ones; transmute() adds new variables and drops existing ones. New variables overwrite existing variables of the same name. Variables can be removed by setting their value to NULL.
starwars %>%
select(name, mass) %>%
mutate(
mass2 = mass * 2,
mass2_squared = mass2 * mass2
)
#> # A tibble: 87 x 4
#> name mass mass2 mass2_squared
#> <chr> <dbl> <dbl> <dbl>
#> 1 Luke Skywalker 77 154 23716
#> 2 C-3PO 75 150 22500
#> 3 R2-D2 32 64 4096
#> 4 Darth Vader 136 272 73984
#> # … with 83 more rows
starwars %>%
transmute(mass2 = mass * 2)
#> # A tibble: 87 x 1
#> mass2
#> <dbl>
#> 1 154
#> 2 150
#> 3 64
#> 4 272
#> # … with 83 more rows
starwars %>%
select(name, height, mass, homeworld) %>%
mutate(
mass = NULL,
height = height * 0.0328084
)
#> # A tibble: 87 x 3
#> name height homeworld
#> <chr> <dbl> <chr>
#> 1 Luke Skywalker 5.64 Tatooine
#> 2 C-3PO 5.48 Tatooine
#> 3 R2-D2 3.15 Naboo
#> 4 Darth Vader 6.63 Tatooine
#> # … with 83 more rows3.6 Extract a single column: pull()
pull() is similar to $. It’s mostly useful because it looks a little nicer in pipes, it can optionally name the output.
mtcars %>% as_tibble()
#> # A tibble: 32 x 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
#> 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
#> 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
#> # … with 28 more rows
mtcars %>% pull(-1)
#> [1] 4 4 1 1 2 1 4 2 2 4 4 3 3 3 4 4 4 1 2 1 1 2 2 4 2 1 2 2 4 6 8 2
mtcars %>% pull(1)
#> [1] 21.0 21.0 22.8 21.4 18.7 18.1 14.3 24.4 22.8 19.2 17.8 16.4 17.3 15.2 10.4
#> [16] 10.4 14.7 32.4 30.4 33.9 21.5 15.5 15.2 13.3 19.2 27.3 26.0 30.4 15.8 19.7
#> [31] 15.0 21.4
mtcars %>% pull(cyl)
#> [1] 6 6 4 6 8 6 8 4 4 6 6 8 8 8 8 8 8 4 4 4 4 8 8 8 8 4 4 4 8 6 8 43.7 Change column order: relocate()
Use relocate() to change column positions.
df <- tibble(a = 1, b = 1, c = 1, d = "a", e = "a", f = "a")
df %>% relocate(f)
#> # A tibble: 1 x 6
#> f a b c d e
#> <chr> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 a 1 1 1 a a
df %>% relocate(a, .after = c)
#> # A tibble: 1 x 6
#> b c a d e f
#> <dbl> <dbl> <dbl> <chr> <chr> <chr>
#> 1 1 1 1 a a a
df %>% relocate(f, .before = b)
#> # A tibble: 1 x 6
#> a f b c d e
#> <dbl> <chr> <dbl> <dbl> <chr> <chr>
#> 1 1 a 1 1 a a3.8 Rename columns: rename() rename_with()
rename() changes the names of individual variables using new_name = old_name syntax; rename_with() renames columns using a function.
starwars %>% rename(home_world = homeworld) %>% relocate(home_world)
#> # A tibble: 87 x 14
#> home_world name height mass hair_color skin_color eye_color birth_year sex
#> <chr> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 Tatooine Luke… 172 77 blond fair blue 19 male
#> 2 Tatooine C-3PO 167 75 <NA> gold yellow 112 none
#> 3 Naboo R2-D2 96 32 <NA> white, bl… red 33 none
#> 4 Tatooine Dart… 202 136 none white yellow 41.9 male
#> # … with 83 more rows, and 5 more variables: gender <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>% rename_with(str_to_upper)
#> # A tibble: 87 x 14
#> NAME HEIGHT MASS HAIR_COLOR SKIN_COLOR EYE_COLOR BIRTH_YEAR SEX GENDER
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: HOMEWORLD <chr>, SPECIES <chr>,
#> # FILMS <list>, VEHICLES <list>, STARSHIPS <list>3.9 Subset rows using their positions: slice() slice_head() slice_tail() slice_min() slice_max() slice_sample()
slice() lets you index rows by their (integer) locations. It allows you to select, remove, and duplicate rows. It is accompanied by a number of helpers for common use cases:
slice_head()andslice_tail()select the first or last rows.slice_sample()randomly selects rows. Use the optionpropto choose a certain proportion of the cases.slice_min()andslice_max()select rows with highest or lowest values of a variable.
starwars %>% slice(5:10)
#> # A tibble: 6 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Leia Or… 150 49 brown light brown 19 fema… femin…
#> 2 Owen La… 178 120 brown, grey light blue 52 male mascu…
#> 3 Beru Wh… 165 75 brown light blue 47 fema… femin…
#> 4 R5-D4 97 32 <NA> white, red red NA none mascu…
#> # … with 2 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>% slice_head(n = 3)
#> # A tibble: 3 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> # … with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>
starwars %>% slice_tail(n = 3)
#> # A tibble: 3 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 BB8 NA NA none none black NA none mascu…
#> 2 Captain… NA NA unknown unknown unknown NA <NA> <NA>
#> 3 Padmé A… 165 45 brown light brown 46 female femin…
#> # … with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>
starwars %>% slice_sample(n = 5)
#> # A tibble: 5 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Watto 137 NA black blue, grey yellow NA male mascu…
#> 2 Mace W… 188 84 none dark brown 72 male mascu…
#> 3 Ben Qu… 163 65 none grey, green… orange NA male mascu…
#> 4 Dud Bo… 94 45 none blue, grey yellow NA male mascu…
#> # … with 1 more row, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>% slice_sample(prop = 0.1)
#> # A tibble: 8 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Ben Qu… 163 65 none grey, gree… orange NA male mascu…
#> 2 Padmé … 165 45 brown light brown 46 fema… femin…
#> 3 Obi-Wa… 182 77 auburn, wh… fair blue-gray 57 male mascu…
#> 4 Qui-Go… 193 89 brown fair blue 92 male mascu…
#> # … with 4 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>% slice_min(height, n = 3)
#> # A tibble: 3 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Yoda 66 17 white green brown 896 male mascu…
#> 2 Ratts Ty… 79 15 none grey, blue unknown NA male mascu…
#> 3 Wicket S… 88 20 brown brown brown 8 male mascu…
#> # … with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>
starwars %>% slice_max(height, n = 3)
#> # A tibble: 3 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Yarael … 264 NA none white yellow NA male mascul…
#> 2 Tarfful 234 136 brown brown blue NA male mascul…
#> 3 Lama Su 229 88 none grey black NA male mascul…
#> # … with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>3.10 Summarise each group to fewer rows: summarise() summarize()
summarise() creates a new data frame. It will have one (or more) rows for each combination of grouping variables; if there are no grouping variables, the output will have a single row summarising all observations in the input. It will contain one column for each grouping variable and one column for each of the summary statistics that you have specified.
summarise() and summarize() are synonyms.
summarise() often work together with group_by().
starwars %>%
summarise(mean_height = mean(height, na.rm = TRUE))
#> # A tibble: 1 x 1
#> mean_height
#> <dbl>
#> 1 174.
starwars %>%
group_by(species) %>%
summarise(mean_height = mean(height, na.rm = TRUE))
#> # A tibble: 38 x 2
#> species mean_height
#> <chr> <dbl>
#> 1 Aleena 79
#> 2 Besalisk 198
#> 3 Cerean 198
#> 4 Chagrian 196
#> # … with 34 more rows3.11 Count observations by group: count() tally() add_count() add_tally()
count() lets you quickly count the unique values of one or more variables, tally() count number of rows. add_count() and add_tally() add a new column with group-wise counts.
starwars %>% count(species)
#> # A tibble: 38 x 2
#> species n
#> <chr> <int>
#> 1 Aleena 1
#> 2 Besalisk 1
#> 3 Cerean 1
#> 4 Chagrian 1
#> # … with 34 more rows
starwars %>% tally()
#> # A tibble: 1 x 1
#> n
#> <int>
#> 1 87
df <- tribble(
~name, ~gender, ~runs,
"Max", "male", 10,
"Sandra", "female", 1,
"Susan", "female", 4
)
df %>% count(gender)
#> # A tibble: 2 x 2
#> gender n
#> <chr> <int>
#> 1 female 2
#> 2 male 1
df %>% count(gender, wt = runs)
#> # A tibble: 2 x 2
#> gender n
#> <chr> <dbl>
#> 1 female 5
#> 2 male 10
df %>% add_count(gender)
#> # A tibble: 3 x 4
#> name gender runs n
#> <chr> <chr> <dbl> <int>
#> 1 Max male 10 1
#> 2 Sandra female 1 2
#> 3 Susan female 4 2
df %>% add_tally()
#> # A tibble: 3 x 4
#> name gender runs n
#> <chr> <chr> <dbl> <int>
#> 1 Max male 10 3
#> 2 Sandra female 1 3
#> 3 Susan female 4 3You can also do it by using group_by() summarise() mutate():
df %>%
group_by(gender) %>%
summarise(n = n()) %>%
ungroup()
#> # A tibble: 2 x 2
#> gender n
#> <chr> <int>
#> 1 female 2
#> 2 male 1
df %>%
group_by(gender) %>%
mutate(n = n()) %>%
ungroup()
#> # A tibble: 3 x 4
#> name gender runs n
#> <chr> <chr> <dbl> <int>
#> 1 Max male 10 1
#> 2 Sandra female 1 2
#> 3 Susan female 4 24 Two(multi) table verbs
4.1 Efficiently bind multiple data frames by row and column: bind_rows() bind_cols()
one <- starwars[1:2, 1:4]
two <- starwars[9:10, 1:4]
bind_rows(one, two)
#> # A tibble: 4 x 4
#> name height mass hair_color
#> <chr> <int> <dbl> <chr>
#> 1 Luke Skywalker 172 77 blond
#> 2 C-3PO 167 75 <NA>
#> 3 Biggs Darklighter 183 84 black
#> 4 Obi-Wan Kenobi 182 77 auburn, white
bind_rows(list(one, two))
#> # A tibble: 4 x 4
#> name height mass hair_color
#> <chr> <int> <dbl> <chr>
#> 1 Luke Skywalker 172 77 blond
#> 2 C-3PO 167 75 <NA>
#> 3 Biggs Darklighter 183 84 black
#> 4 Obi-Wan Kenobi 182 77 auburn, white
bind_rows(list(one, two), .id = "id")
#> # A tibble: 4 x 5
#> id name height mass hair_color
#> <chr> <chr> <int> <dbl> <chr>
#> 1 1 Luke Skywalker 172 77 blond
#> 2 1 C-3PO 167 75 <NA>
#> 3 2 Biggs Darklighter 183 84 black
#> 4 2 Obi-Wan Kenobi 182 77 auburn, white
bind_rows("group 1" = one, "group 2" = two, .id = "groups")
#> # A tibble: 4 x 5
#> groups name height mass hair_color
#> <chr> <chr> <int> <dbl> <chr>
#> 1 group 1 Luke Skywalker 172 77 blond
#> 2 group 1 C-3PO 167 75 <NA>
#> 3 group 2 Biggs Darklighter 183 84 black
#> 4 group 2 Obi-Wan Kenobi 182 77 auburn, white
bind_cols(one, two)
#> New names:
#> * name -> name...1
#> * height -> height...2
#> * mass -> mass...3
#> * hair_color -> hair_color...4
#> * name -> name...5
#> * ...
#> # A tibble: 2 x 8
#> name...1 height...2 mass...3 hair_color...4 name...5 height...6 mass...7
#> <chr> <int> <dbl> <chr> <chr> <int> <dbl>
#> 1 Luke Skywa… 172 77 blond Biggs Dark… 183 84
#> 2 C-3PO 167 75 <NA> Obi-Wan Ke… 182 77
#> # … with 1 more variable: hair_color...8 <chr>4.2 Set operations: intersect() union() union_all() setdiff()
Set operation functions expect the x and y inputs to have the same variables, and treat the observations like sets:
intersect(x, y): return only observations in bothxandyunion(x, y): return unique observations inxandyunion_all(x, y): return all observations inxandysetdiff(x, y): return observations inx, but not iny.
Beware that intersect(), union() and setdiff() remove duplicates.
(df1 <- tibble(x = 1:2, y = c(1L, 1L)))
#> # A tibble: 2 x 2
#> x y
#> <int> <int>
#> 1 1 1
#> 2 2 1
(df2 <- tibble(x = 1:2, y = 1:2))
#> # A tibble: 2 x 2
#> x y
#> <int> <int>
#> 1 1 1
#> 2 2 2
intersect(df1, df2)
#> # A tibble: 1 x 2
#> x y
#> <int> <int>
#> 1 1 1
union(df1, df2)
#> # A tibble: 3 x 2
#> x y
#> <int> <int>
#> 1 1 1
#> 2 2 1
#> 3 2 2
union_all(df1, df2)
#> # A tibble: 4 x 2
#> x y
#> <int> <int>
#> 1 1 1
#> 2 2 1
#> 3 1 1
#> 4 2 2
setdiff(df1, df2)
#> # A tibble: 1 x 2
#> x y
#> <int> <int>
#> 1 2 1
setdiff(df2, df1)
#> # A tibble: 1 x 2
#> x y
#> <int> <int>
#> 1 2 24.3 Mutating joins: inner_join() left_join() right_join() full_join()
The mutating joins add columns from y to x, matching rows based on the keys:
inner_join(): includes all rows inxandy.left_join(): includes all rows inx.right_join(): includes all rows iny.full_join(): includes all rows inxory.
If a row in x matches multiple rows in y, all the rows in y will be returned once for each matching row in x.
band_members
#> # A tibble: 3 x 2
#> name band
#> <chr> <chr>
#> 1 Mick Stones
#> 2 John Beatles
#> 3 Paul Beatles
band_instruments
#> # A tibble: 3 x 2
#> name plays
#> <chr> <chr>
#> 1 John guitar
#> 2 Paul bass
#> 3 Keith guitar
band_members %>% inner_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 2 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 John Beatles guitar
#> 2 Paul Beatles bass
band_members %>% left_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 3 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 Mick Stones <NA>
#> 2 John Beatles guitar
#> 3 Paul Beatles bass
band_members %>% right_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 3 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 John Beatles guitar
#> 2 Paul Beatles bass
#> 3 Keith <NA> guitar
band_members %>% full_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 4 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 Mick Stones <NA>
#> 2 John Beatles guitar
#> 3 Paul Beatles bass
#> 4 Keith <NA> guitarThe value of these mutating join functions are an object of the same type as x. The order of the rows and columns of x is preserved as much as possible. The output has the following properties:
For
inner_join(), a subset ofxrows. Forleft_join(), allxrows. Forright_join(), a subset ofxrows, followed by unmatchedyrows. Forfull_join(), allxrows, followed by unmatchedyrows.For all joins, rows will be duplicated if one or more rows in
xmatches multiple rows iny.Output columns include all
xcolumns and allycolumns. If columns inxandyhave the same name (and aren’t included inby),suffixes are added to disambiguate.Output columns included in
byare coerced to common type acrossxandy.Groups are taken from
x.
band_members
#> # A tibble: 3 x 2
#> name band
#> <chr> <chr>
#> 1 Mick Stones
#> 2 John Beatles
#> 3 Paul Beatles
band_instruments
#> # A tibble: 3 x 2
#> name plays
#> <chr> <chr>
#> 1 John guitar
#> 2 Paul bass
#> 3 Keith guitar
band_instruments2
#> # A tibble: 3 x 2
#> artist plays
#> <chr> <chr>
#> 1 John guitar
#> 2 Paul bass
#> 3 Keith guitar
band_members %>% inner_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 2 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 John Beatles guitar
#> 2 Paul Beatles bass
band_members %>% inner_join(band_instruments, by = "name")
#> # A tibble: 2 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 John Beatles guitar
#> 2 Paul Beatles bass
band_members %>% inner_join(band_instruments2, by = c("name" = "artist"))
#> # A tibble: 2 x 3
#> name band plays
#> <chr> <chr> <chr>
#> 1 John Beatles guitar
#> 2 Paul Beatles bass
band_members %>%
inner_join(band_instruments2, by = c("name" = "artist"), keep = TRUE)
#> # A tibble: 2 x 4
#> name band artist plays
#> <chr> <chr> <chr> <chr>
#> 1 John Beatles John guitar
#> 2 Paul Beatles Paul bass
band_members %>%
inner_join(band_instruments, by = "name", keep = TRUE)
#> # A tibble: 2 x 4
#> name.x band name.y plays
#> <chr> <chr> <chr> <chr>
#> 1 John Beatles John guitar
#> 2 Paul Beatles Paul bass
band_members %>%
inner_join(band_instruments, by = "name", keep = TRUE, suffix = c('_members', '_instruments'))
#> # A tibble: 2 x 4
#> name_members band name_instruments plays
#> <chr> <chr> <chr> <chr>
#> 1 John Beatles John guitar
#> 2 Paul Beatles Paul bass4.4 Filtering joins: semi_join() anti_join()
Filtering joins filter rows from x based on the presence or absence of matches in y:
semi_join()return all rows fromxwith a match iny.anti_join()return all rows fromxwithout a match iny.
band_members
#> # A tibble: 3 x 2
#> name band
#> <chr> <chr>
#> 1 Mick Stones
#> 2 John Beatles
#> 3 Paul Beatles
band_instruments
#> # A tibble: 3 x 2
#> name plays
#> <chr> <chr>
#> 1 John guitar
#> 2 Paul bass
#> 3 Keith guitar
band_members %>% semi_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 2 x 2
#> name band
#> <chr> <chr>
#> 1 John Beatles
#> 2 Paul Beatles
band_members %>% anti_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 1 x 2
#> name band
#> <chr> <chr>
#> 1 Mick StonesFiltering joins perform like filter() rather than mutating joins.
band_members %>% filter(name %in% band_instruments$name)
#> # A tibble: 2 x 2
#> name band
#> <chr> <chr>
#> 1 John Beatles
#> 2 Paul Beatles
band_members %>% filter(!(name %in% band_instruments$name))
#> # A tibble: 1 x 2
#> name band
#> <chr> <chr>
#> 1 Mick Stones4.5 Nest join: nest_join()
nest_join() returns all rows and columns in x with a new nested-df column that contains all matches from y. When there is no match, the list column is a 0-row tibble.
band_members %>% nest_join(band_instruments)
#> Joining, by = "name"
#> # A tibble: 3 x 3
#> name band band_instruments
#> <chr> <chr> <list>
#> 1 Mick Stones <tibble[,1] [0 × 1]>
#> 2 John Beatles <tibble[,1] [1 × 1]>
#> 3 Paul Beatles <tibble[,1] [1 × 1]>In some sense, a nest_join() is the most fundamental join since you can recreate the other joins from it:
inner_join()is anest_join()plustidyr::unnest(),left_join()is anest_join()plusunnest(.drop = FALSE),semi_join()is anest_join()plus afilter()where you check that every element of data has at least one row,anti_join()is anest_join()plus afilter()where you check every element has zero rows.
5 Tidy evaluation
Most dplyr verbs use tidy evaluation in some way. Tidy evaluation is a special type of non-standard evaluation used throughout the tidyverse. There are two basic forms found in dplyr:
arrange(),count(),add_count(),distinct(),filter(),group_by(),mutate(),transmute(), andsummarise()use data masking so that you can use data variables as if they were variables in the environment (i.e. you writemy_variablenotdf$my_variable).across(),if_any(),if_all(),relocate(),rename(),select(), andpull()use tidy selection so you can easily choose variables based on their position, name, or type (e.g.starts_with("x")oris.numeric).
To determine whether a function argument uses data masking or tidy selection, look at the documentation: in the arguments list, you’ll see <data-masking> or <tidy-select>.
5.1 Data masking
Data masking makes data manipulation faster because it requires less typing. In most (but not all) base R functions you need to refer to variables with $, leading to code that repeats the name of the data frame many times:
starwars[starwars$homeworld == "Naboo" & starwars$species == "Human", ]The dplyr equivalent of this code is more concise because data masking allows you to need to type starwars once:
starwars %>% filter(homeworld == "Naboo", species == "Human")5.1.1 Data- and env-variables
The key idea behind data masking is that it blurs the line between the two different meanings of the word “variable”:
env-variables are “programming” variables that live in an environment. They are usually created with
<-.data-variables are “statistical” variables that live in a data frame. They usually come from data files (e.g.
.csv,.xls), or are created manipulating existing variables.
To make those definitions a little more concrete, take this piece of code:
df <- tibble(x = runif(3), y = runif(3))
df$x
#> [1] 0.63847545 0.06882137 0.57590635It creates a env-variable, df, that contains two data-variables, x and y. Then it extracts the data-variable x out of the env-variable df using $.
This blurring of the meaning of “variable” is a really nice feature for interactive data analysis because it allows you to refer to data-vars as is, without any prefix. And this seems to be fairly intuitive since many newer R users will attempt to write starwars[homeworld == "Naboo" & species == "Human", ].
5.1.2 Call data- or env-variables straightforwardly
Data masking is convenient in most cases, but it would be dangerous if you want to use both data-variables and env-variables in the same time. They might own the same variable name so it’s confused which variable has been used.
Let’s begin with this code block
df <- tibble(x = 1:3)
min_x <- 2
df %>% filter(x >= min_x)
#> # A tibble: 2 x 1
#> x
#> <int>
#> 1 2
#> 2 3It works well, but what would happen if there is a data-variable min_x in df?
df <- tibble(x = 1:3, min_x = 0)
min_x <- 2
df %>% filter(x >= min_x)
#> # A tibble: 3 x 2
#> x min_x
#> <int> <dbl>
#> 1 1 0
#> 2 2 0
#> 3 3 0There are no rows been dropped now. So it would be dangerous if you want to use both data-variables and env-variables in the same time, especially working with user-supplied data.
In this case, you need use .data and .env to declare witch variable you want to use.
df %>% filter(.data$x >= .env$min_x)
#> # A tibble: 2 x 2
#> x min_x
#> <int> <dbl>
#> 1 2 0
#> 2 3 05.2 Tidy selection
Data masking makes it easy to compute on values within a dataset. Tidy selection is a complementary tool that makes it easy to work with the (group of) columns of a dataset. It provides a concise dialect of R for selecting variables based on their names or properties.
5.2.1 Overview of selection features
Tidyverse selections implement a dialect of R where operators make it easy to select variables:
:for selecting a range of consecutive variables.!for taking the complement of a set of variables.&and|for selecting the intersection or the union of two sets of variables.c()for combining selections.iris <- iris %>% as_tibble() starwars %>% select(name:mass) #> # A tibble: 87 x 3 #> name height mass #> <chr> <int> <dbl> #> 1 Luke Skywalker 172 77 #> 2 C-3PO 167 75 #> 3 R2-D2 96 32 #> 4 Darth Vader 202 136 #> # … with 83 more rows starwars %>% select(!(name:mass)) #> # A tibble: 87 x 11 #> hair_color skin_color eye_color birth_year sex gender homeworld species #> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> #> 1 blond fair blue 19 male masculine Tatooine Human #> 2 <NA> gold yellow 112 none masculine Tatooine Droid #> 3 <NA> white, blue red 33 none masculine Naboo Droid #> 4 none white yellow 41.9 male masculine Tatooine Human #> # … with 83 more rows, and 3 more variables: films <list>, vehicles <list>, #> # starships <list> iris %>% select(starts_with("Petal") & ends_with("Width")) #> # A tibble: 150 x 1 #> Petal.Width #> <dbl> #> 1 0.2 #> 2 0.2 #> 3 0.2 #> 4 0.2 #> # … with 146 more rows iris %>% select(starts_with("Petal") | ends_with("Width")) #> # A tibble: 150 x 3 #> Petal.Length Petal.Width Sepal.Width #> <dbl> <dbl> <dbl> #> 1 1.4 0.2 3.5 #> 2 1.4 0.2 3 #> 3 1.3 0.2 3.2 #> 4 1.5 0.2 3.1 #> # … with 146 more rows iris %>% select(c(starts_with("Petal"), Species)) #> # A tibble: 150 x 3 #> Petal.Length Petal.Width Species #> <dbl> <dbl> <fct> #> 1 1.4 0.2 setosa #> 2 1.4 0.2 setosa #> 3 1.3 0.2 setosa #> 4 1.5 0.2 setosa #> # … with 146 more rows
In addition, you can use selection helpers. Some helpers select specific columns:
everything(): Matches all variables.last_col(): Select last variable, possibly with an offset.iris %>% select(everything()) #> # A tibble: 150 x 5 #> Sepal.Length Sepal.Width Petal.Length Petal.Width Species #> <dbl> <dbl> <dbl> <dbl> <fct> #> 1 5.1 3.5 1.4 0.2 setosa #> 2 4.9 3 1.4 0.2 setosa #> 3 4.7 3.2 1.3 0.2 setosa #> 4 4.6 3.1 1.5 0.2 setosa #> # … with 146 more rows iris %>% select(last_col()) #> # A tibble: 150 x 1 #> Species #> <fct> #> 1 setosa #> 2 setosa #> 3 setosa #> 4 setosa #> # … with 146 more rows iris %>% select(last_col(0:2)) #> # A tibble: 150 x 3 #> Species Petal.Width Petal.Length #> <fct> <dbl> <dbl> #> 1 setosa 0.2 1.4 #> 2 setosa 0.2 1.4 #> 3 setosa 0.2 1.3 #> 4 setosa 0.2 1.5 #> # … with 146 more rows
These helpers select variables by matching patterns in their names:
starts_with(): Starts with a prefix.ends_with(): Ends with a suffix.contains(): Contains a literal string.matches(): Matches a regular expression.num_range(): Matches a numerical range like x01, x02, x03.iris %>% select(starts_with("Sepal")) #> # A tibble: 150 x 2 #> Sepal.Length Sepal.Width #> <dbl> <dbl> #> 1 5.1 3.5 #> 2 4.9 3 #> 3 4.7 3.2 #> 4 4.6 3.1 #> # … with 146 more rows iris %>% select(ends_with("Width")) #> # A tibble: 150 x 2 #> Sepal.Width Petal.Width #> <dbl> <dbl> #> 1 3.5 0.2 #> 2 3 0.2 #> 3 3.2 0.2 #> 4 3.1 0.2 #> # … with 146 more rows iris %>% select(starts_with(c("Petal", "Sepal"))) #> # A tibble: 150 x 4 #> Petal.Length Petal.Width Sepal.Length Sepal.Width #> <dbl> <dbl> <dbl> <dbl> #> 1 1.4 0.2 5.1 3.5 #> 2 1.4 0.2 4.9 3 #> 3 1.3 0.2 4.7 3.2 #> 4 1.5 0.2 4.6 3.1 #> # … with 146 more rows iris %>% select(ends_with(c("Width", "Length"))) #> # A tibble: 150 x 4 #> Sepal.Width Petal.Width Sepal.Length Petal.Length #> <dbl> <dbl> <dbl> <dbl> #> 1 3.5 0.2 5.1 1.4 #> 2 3 0.2 4.9 1.4 #> 3 3.2 0.2 4.7 1.3 #> 4 3.1 0.2 4.6 1.5 #> # … with 146 more rows iris %>% select(contains("al")) #> # A tibble: 150 x 4 #> Sepal.Length Sepal.Width Petal.Length Petal.Width #> <dbl> <dbl> <dbl> <dbl> #> 1 5.1 3.5 1.4 0.2 #> 2 4.9 3 1.4 0.2 #> 3 4.7 3.2 1.3 0.2 #> 4 4.6 3.1 1.5 0.2 #> # … with 146 more rows iris %>% select(matches("[pt]al")) #> # A tibble: 150 x 4 #> Sepal.Length Sepal.Width Petal.Length Petal.Width #> <dbl> <dbl> <dbl> <dbl> #> 1 5.1 3.5 1.4 0.2 #> 2 4.9 3 1.4 0.2 #> 3 4.7 3.2 1.3 0.2 #> 4 4.6 3.1 1.5 0.2 #> # … with 146 more rows billboard %>% select(starts_with("wk")) #> # A tibble: 317 x 76 #> wk1 wk2 wk3 wk4 wk5 wk6 wk7 wk8 wk9 wk10 wk11 wk12 wk13 #> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 87 82 72 77 87 94 99 NA NA NA NA NA NA #> 2 91 87 92 NA NA NA NA NA NA NA NA NA NA #> 3 81 70 68 67 66 57 54 53 51 51 51 51 47 #> 4 76 76 72 69 67 65 55 59 62 61 61 59 61 #> # … with 313 more rows, and 63 more variables: wk14 <dbl>, wk15 <dbl>, #> # wk16 <dbl>, wk17 <dbl>, wk18 <dbl>, wk19 <dbl>, wk20 <dbl>, wk21 <dbl>, #> # wk22 <dbl>, wk23 <dbl>, wk24 <dbl>, wk25 <dbl>, wk26 <dbl>, wk27 <dbl>, #> # wk28 <dbl>, wk29 <dbl>, wk30 <dbl>, wk31 <dbl>, wk32 <dbl>, wk33 <dbl>, #> # wk34 <dbl>, wk35 <dbl>, wk36 <dbl>, wk37 <dbl>, wk38 <dbl>, wk39 <dbl>, #> # wk40 <dbl>, wk41 <dbl>, wk42 <dbl>, wk43 <dbl>, wk44 <dbl>, wk45 <dbl>, #> # wk46 <dbl>, wk47 <dbl>, wk48 <dbl>, wk49 <dbl>, wk50 <dbl>, wk51 <dbl>, #> # wk52 <dbl>, wk53 <dbl>, wk54 <dbl>, wk55 <dbl>, wk56 <dbl>, wk57 <dbl>, #> # wk58 <dbl>, wk59 <dbl>, wk60 <dbl>, wk61 <dbl>, wk62 <dbl>, wk63 <dbl>, #> # wk64 <dbl>, wk65 <dbl>, wk66 <lgl>, wk67 <lgl>, wk68 <lgl>, wk69 <lgl>, #> # wk70 <lgl>, wk71 <lgl>, wk72 <lgl>, wk73 <lgl>, wk74 <lgl>, wk75 <lgl>, #> # wk76 <lgl> billboard %>% select(num_range("wk", 10:15)) #> # A tibble: 317 x 6 #> wk10 wk11 wk12 wk13 wk14 wk15 #> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 NA NA NA NA NA NA #> 2 NA NA NA NA NA NA #> 3 51 51 51 47 44 38 #> 4 61 61 59 61 66 72 #> # … with 313 more rows
These helpers select variables from a character vector:
all_of(): Matches variable names in a character vector. All names must be present, otherwise an out-of-bounds error is thrown.any_of(): Same asall_of(), except that no error is thrown for names that don’t exist.vars <- c("Sepal.Length", "Sepal.Width") iris %>% select(all_of(vars)) #> # A tibble: 150 x 2 #> Sepal.Length Sepal.Width #> <dbl> <dbl> #> 1 5.1 3.5 #> 2 4.9 3 #> 3 4.7 3.2 #> 4 4.6 3.1 #> # … with 146 more rows starwars %>% select(all_of(vars)) #> Error: Can't subset columns that don't exist. #> x Columns `Sepal.Length` and `Sepal.Width` don't exist. starwars %>% select(any_of(vars)) #> # A tibble: 87 x 0
This helper selects variables with a function:
where(): Applies a function to all variables and selects those for which the function returnsTRUE.iris %>% select(where(is.factor)) #> # A tibble: 150 x 1 #> Species #> <fct> #> 1 setosa #> 2 setosa #> 3 setosa #> 4 setosa #> # … with 146 more rows iris %>% select(where(is.numeric)) #> # A tibble: 150 x 4 #> Sepal.Length Sepal.Width Petal.Length Petal.Width #> <dbl> <dbl> <dbl> <dbl> #> 1 5.1 3.5 1.4 0.2 #> 2 4.9 3 1.4 0.2 #> 3 4.7 3.2 1.3 0.2 #> 4 4.6 3.1 1.5 0.2 #> # … with 146 more rows iris %>% select(where(~ is.numeric(.x))) #> # A tibble: 150 x 4 #> Sepal.Length Sepal.Width Petal.Length Petal.Width #> <dbl> <dbl> <dbl> <dbl> #> 1 5.1 3.5 1.4 0.2 #> 2 4.9 3 1.4 0.2 #> 3 4.7 3.2 1.3 0.2 #> 4 4.6 3.1 1.5 0.2 #> # … with 146 more rows iris %>% select(where(~ is.numeric(.x) && mean(.x) > 3.5)) #> # A tibble: 150 x 2 #> Sepal.Length Petal.Length #> <dbl> <dbl> #> 1 5.1 1.4 #> 2 4.9 1.4 #> 3 4.7 1.3 #> 4 4.6 1.5 #> # … with 146 more rows
5.3 Column-wise operations
You can also use ‘tidy-select’ semantics in ‘data-masking’ functions, with the help of across() if_any() if_all(). They make it easy to apply the same transformation to multiple columns.
5.3.1 Basic usage
across() has two primary arguments:
The first argument,
.cols, selects the columns you want to operate on. It uses tidy selection (likeselect()) so you can pick variables by position, name, and type.The second argument,
.fns, is a function or list of functions to apply to each column. This can also be a purrr style formula (or list of formulas) like~ .x / 2. (This argument is optional, and you can omit it if you just want to get the underlying data.)
5.3.2 Omit .fns
You can use ‘tidy-select’ semantics to select columns and then do future work based on the verb itself by omitting .fns.
starwars %>% distinct(across(contains("color")))
#> # A tibble: 67 x 3
#> hair_color skin_color eye_color
#> <chr> <chr> <chr>
#> 1 blond fair blue
#> 2 <NA> gold yellow
#> 3 <NA> white, blue red
#> 4 none white yellow
#> # … with 63 more rows
starwars %>% count(across(contains("color")), sort = TRUE)
#> # A tibble: 67 x 4
#> hair_color skin_color eye_color n
#> <chr> <chr> <chr> <int>
#> 1 brown light brown 6
#> 2 brown fair blue 4
#> 3 none grey black 4
#> 4 black dark brown 3
#> # … with 63 more rows
iris %>%
as_tibble() %>%
mutate(sum_length = rowSums(across(ends_with('Length'))))
#> # A tibble: 150 x 6
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species sum_length
#> <dbl> <dbl> <dbl> <dbl> <fct> <dbl>
#> 1 5.1 3.5 1.4 0.2 setosa 6.5
#> 2 4.9 3 1.4 0.2 setosa 6.3
#> 3 4.7 3.2 1.3 0.2 setosa 6
#> 4 4.6 3.1 1.5 0.2 setosa 6.1
#> # … with 146 more rows5.3.3 Usage of .fns
You can use .fns to generate new column (usually used in combination with summarise() and mutate()).
starwars %>%
summarise(across(where(is.character), ~ length(unique(.x))))
#> # A tibble: 1 x 8
#> name hair_color skin_color eye_color sex gender homeworld species
#> <int> <int> <int> <int> <int> <int> <int> <int>
#> 1 87 13 31 15 5 3 49 38
starwars %>%
summarise(across(where(is.character), ~ length(unique(.x)), .names = 'count_{.col}'))
#> # A tibble: 1 x 8
#> count_name count_hair_color count_skin_color count_eye_color count_sex
#> <int> <int> <int> <int> <int>
#> 1 87 13 31 15 5
#> # … with 3 more variables: count_gender <int>, count_homeworld <int>,
#> # count_species <int>
starwars %>%
mutate(across(!where(is.list), as.character))
#> # A tibble: 87 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
starwars %>%
mutate(across(!where(is.list), as.character)) %>%
summarise(across(where(is.character), ~ max(0, str_length(.x), na.rm = T)))
#> # A tibble: 1 x 11
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 3 4 13 19 13 4 14 9
#> # … with 2 more variables: homeworld <dbl>, species <dbl>5.3.4 Multiple functions
You can transform each variable with more than one function by supplying a named list of functions or lambda functions in the second argument:
min_max <- list(
min = ~min(.x, na.rm = TRUE),
max = ~max(.x, na.rm = TRUE)
)
starwars %>% summarise(across(where(is.numeric), min_max))
#> # A tibble: 1 x 6
#> height_min height_max mass_min mass_max birth_year_min birth_year_max
#> <int> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 66 264 15 1358 8 896Control how the names are created with the .names argument which takes a glue spec:
starwars %>% summarise(across(where(is.numeric), min_max, .names = "{.fn}.{.col}"))
#> # A tibble: 1 x 6
#> min.height max.height min.mass max.mass min.birth_year max.birth_year
#> <int> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 66 264 15 1358 8 896We can work around this by combining both calls to across() into a single expression that returns a tibble:
starwars %>% summarise(
tibble(
across(where(is.numeric), ~min(.x, na.rm = TRUE), .names = "min_{.col}"),
across(where(is.numeric), ~max(.x, na.rm = TRUE), .names = "max_{.col}")
)
)
#> # A tibble: 1 x 6
#> min_height min_mass min_birth_year max_height max_mass max_birth_year
#> <int> <dbl> <dbl> <int> <dbl> <dbl>
#> 1 66 15 8 264 1358 8965.3.5 Current column
If you need to, you can access the name of the “current” column inside by calling cur_column():
df <- tibble(x = 1:3, y = 3:5, z = 5:7)
mult <- list(x = 1, y = 10, z = 100)
df %>% mutate(across(everything(), ~ .x * mult[[cur_column()]]))
#> # A tibble: 3 x 3
#> x y z
#> <dbl> <dbl> <dbl>
#> 1 1 30 500
#> 2 2 40 600
#> 3 3 50 700
df %>% mutate(across(everything(), ~ str_c(cur_column(), .x, sep = ': ')))
#> # A tibble: 3 x 3
#> x y z
#> <chr> <chr> <chr>
#> 1 x: 1 y: 3 z: 5
#> 2 x: 2 y: 4 z: 6
#> 3 x: 3 y: 5 z: 75.3.6 filter()
We cannot directly use across() in filter() because we need an extra step to combine the results. To that end, filter() has two special purpose companion functions (newly in dplyr v1.0.4):
if_any()keeps the rows where the predicate is true for at least one selected column:starwars %>% filter(if_any(everything(), ~ !is.na(.x))) #> # A tibble: 87 x 14 #> name height mass hair_color skin_color eye_color birth_year sex gender #> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> #> 1 Luke Sk… 172 77 blond fair blue 19 male mascu… #> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu… #> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu… #> 4 Darth V… 202 136 none white yellow 41.9 male mascu… #> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>, #> # films <list>, vehicles <list>, starships <list>if_all()keeps the rows where the predicate is true for all selected columns:starwars %>% filter(if_all(everything(), ~ !is.na(.x))) #> # A tibble: 29 x 14 #> name height mass hair_color skin_color eye_color birth_year sex gender #> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> #> 1 Luke Sk… 172 77 blond fair blue 19 male mascu… #> 2 Darth V… 202 136 none white yellow 41.9 male mascu… #> 3 Leia Or… 150 49 brown light brown 19 fema… femin… #> 4 Owen La… 178 120 brown, grey light blue 52 male mascu… #> # … with 25 more rows, and 5 more variables: homeworld <chr>, species <chr>, #> # films <list>, vehicles <list>, starships <list>
You can use across() to do the same things with some extra steps:
iris %>%
as_tibble() %>%
filter(if_any(ends_with("Width"), ~ . > 4))
#> # A tibble: 3 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.7 4.4 1.5 0.4 setosa
#> 2 5.2 4.1 1.5 0.1 setosa
#> 3 5.5 4.2 1.4 0.2 setosa
iris %>%
as_tibble() %>%
filter(rowSums(across(ends_with("Width"), ~ . > 4)) > 0)
#> # A tibble: 3 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 5.7 4.4 1.5 0.4 setosa
#> 2 5.2 4.1 1.5 0.1 setosa
#> 3 5.5 4.2 1.4 0.2 setosa
iris %>%
as_tibble() %>%
filter(if_all(ends_with("Width"), ~ . > 2))
#> # A tibble: 23 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 6.3 3.3 6 2.5 virginica
#> 2 7.1 3 5.9 2.1 virginica
#> 3 6.5 3 5.8 2.2 virginica
#> 4 7.6 3 6.6 2.1 virginica
#> # … with 19 more rows
iris %>%
as_tibble() %>%
filter(rowSums(!across(ends_with("Width"), ~ . > 2)) == 0)
#> # A tibble: 23 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <fct>
#> 1 6.3 3.3 6 2.5 virginica
#> 2 7.1 3 5.9 2.1 virginica
#> 3 6.5 3 5.8 2.2 virginica
#> 4 7.6 3 6.6 2.1 virginica
#> # … with 19 more rows5.4 Programming with dplyr
Data masking and tidy selection make interactive data exploration fast and fluid, but they add some new challenges when you attempt to use them indirectly such as in a for loop or a function. If you want to write some functions with the technique of <data-masking> and <tidy-select>, you can get help in ?dplyr_data_masking ?dplyr_tidy_select and vignette("programming", package = "dplyr").
6 Grouped data
Most data operations are done on groups defined by variables. group_by() takes an existing tbl and converts it into a grouped tbl where operations are performed “by group”. ungroup() removes grouping.
by_species <- starwars %>% group_by(species)
by_sex_gender <- starwars %>% group_by(sex, gender)You can see the grouping when you print the data:
by_species
#> # A tibble: 87 x 14
#> # Groups: species [38]
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>
by_sex_gender
#> # A tibble: 87 x 14
#> # Groups: sex, gender [6]
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>If you apply group_by() to an already grouped dataset, will overwrite the existing grouping variables. For example, the following code groups by homeworld instead of species:
by_species %>% group_by(homeworld)
#> # A tibble: 87 x 14
#> # Groups: homeworld [49]
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>To augment the grouping, using .add = TRUE. For example, the following code groups by species and homeworld:
by_species %>% group_by(homeworld, .add = TRUE)
#> # A tibble: 87 x 14
#> # Groups: species, homeworld [58]
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>To remove all grouping variables, use ungroup():
by_species %>% ungroup()
#> # A tibble: 87 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>You can also choose to selectively ungroup by listing the variables you want to remove:
by_sex_gender %>% ungroup(sex)
#> # A tibble: 87 x 14
#> # Groups: gender [3]
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk… 172 77 blond fair blue 19 male mascu…
#> 2 C-3PO 167 75 <NA> gold yellow 112 none mascu…
#> 3 R2-D2 96 32 <NA> white, blue red 33 none mascu…
#> 4 Darth V… 202 136 none white yellow 41.9 male mascu…
#> # … with 83 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>6.1 How grouping affects the main dplyr verbs
6.1.1 summarise()
summarise() computes a summary for each group. It starts from grouping variables, adding summary variables to the right hand side:
by_species %>%
summarise(
n = n(),
height = mean(height, na.rm = TRUE)
)
#> # A tibble: 38 x 3
#> species n height
#> <chr> <int> <dbl>
#> 1 Aleena 1 79
#> 2 Besalisk 1 198
#> 3 Cerean 1 198
#> 4 Chagrian 1 196
#> # … with 34 more rows6.1.2 tally() and add_tally()
tally() and add_tally() count number of rows by groups.
by_species %>% tally()
#> # A tibble: 38 x 2
#> species n
#> <chr> <int>
#> 1 Aleena 1
#> 2 Besalisk 1
#> 3 Cerean 1
#> 4 Chagrian 1
#> # … with 34 more rows
by_sex_gender %>% tally()
#> # A tibble: 6 x 3
#> # Groups: sex [5]
#> sex gender n
#> <chr> <chr> <int>
#> 1 female feminine 16
#> 2 hermaphroditic masculine 1
#> 3 male masculine 60
#> 4 none feminine 1
#> # … with 2 more rows6.1.3 select(), rename(), and relocate()
rename() and relocate() behave identically with grouped and ungrouped data because they only affect the name or position of existing columns. Grouped select() is almost identical to ungrouped select, except that it always includes the grouping variables:
by_species %>% select(mass)
#> Adding missing grouping variables: `species`
#> # A tibble: 87 x 2
#> # Groups: species [38]
#> species mass
#> <chr> <dbl>
#> 1 Human 77
#> 2 Droid 75
#> 3 Droid 32
#> 4 Human 136
#> # … with 83 more rows6.1.4 arrange()
Grouped arrange() is the same as ungrouped arrange()
6.1.5 distinct()
The same behavior applies for grouped data frames, except that the grouping variables are always included:
by_species %>% distinct(mass)
#> # A tibble: 62 x 2
#> # Groups: species [38]
#> mass species
#> <dbl> <chr>
#> 1 77 Human
#> 2 75 Droid
#> 3 32 Droid
#> 4 136 Human
#> # … with 58 more rows6.1.6 mutate() and transmute()
In simple cases with vectorised functions, grouped and ungrouped mutate() give the same results. They differ when used with summary (or window) functions:
by_species %>%
select(mass) %>%
mutate(mean_mass = mean(mass, na.rm = TRUE))
#> Adding missing grouping variables: `species`
#> # A tibble: 87 x 3
#> # Groups: species [38]
#> species mass mean_mass
#> <chr> <dbl> <dbl>
#> 1 Human 77 82.8
#> 2 Droid 75 69.8
#> 3 Droid 32 69.8
#> 4 Human 136 82.8
#> # … with 83 more rows
by_species %>%
select(mass) %>%
mutate(lag_mass = lag(mass))
#> Adding missing grouping variables: `species`
#> # A tibble: 87 x 3
#> # Groups: species [38]
#> species mass lag_mass
#> <chr> <dbl> <dbl>
#> 1 Human 77 NA
#> 2 Droid 75 NA
#> 3 Droid 32 75
#> 4 Human 136 77
#> # … with 83 more rows6.1.7 filter()
A grouped filter() effectively does a mutate() to generate a logical variable, and then only keeps the rows where the variable is TRUE. This means that grouped filters can be used with summary functions. For example, we can find the tallest character of each species:
by_species %>%
select(name, species, height) %>%
filter(height == max(height))
#> # A tibble: 35 x 3
#> # Groups: species [35]
#> name species height
#> <chr> <chr> <int>
#> 1 Greedo Rodian 173
#> 2 Jabba Desilijic Tiure Hutt 175
#> 3 Yoda Yoda's species 66
#> 4 Bossk Trandoshan 190
#> # … with 31 more rowsYou can also use filter() to remove entire groups. For example, the following code eliminates all groups that only have a single member:
by_species %>%
filter(n() != 1) %>%
tally()
#> # A tibble: 9 x 2
#> species n
#> <chr> <int>
#> 1 Droid 6
#> 2 Gungan 3
#> 3 Human 35
#> 4 Kaminoan 2
#> # … with 5 more rows6.1.8 slice() and friends
slice() and friends (slice_head(), slice_tail(), slice_sample(), slice_min() and slice_max()) select rows within a group. For example, we can select the first observation within each species:
by_species %>%
relocate(species) %>%
slice(1)
#> # A tibble: 38 x 14
#> # Groups: species [38]
#> species name height mass hair_color skin_color eye_color birth_year sex
#> <chr> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
#> 1 Aleena Ratts … 79 15 none grey, blue unknown NA male
#> 2 Besalisk Dexter… 198 102 none brown yellow NA male
#> 3 Cerean Ki-Adi… 198 82 white pale yellow 92 male
#> 4 Chagrian Mas Am… 196 NA none blue blue NA male
#> # … with 34 more rows, and 5 more variables: gender <chr>, homeworld <chr>,
#> # films <list>, vehicles <list>, starships <list>6.2 Information of grouped data
6.2.1 Grouping metadata
You can see underlying group data with group_keys(). It has one row for each group and one column for each grouping variable:
by_species %>% group_keys()
#> # A tibble: 38 x 1
#> species
#> <chr>
#> 1 Aleena
#> 2 Besalisk
#> 3 Cerean
#> 4 Chagrian
#> # … with 34 more rows
by_sex_gender %>% group_keys()
#> # A tibble: 6 x 2
#> sex gender
#> <chr> <chr>
#> 1 female feminine
#> 2 hermaphroditic masculine
#> 3 male masculine
#> 4 none feminine
#> # … with 2 more rowsYou can see which group each row belongs to with group_indices():
by_sex_gender %>% group_indices()
#> [1] 3 5 5 3 1 3 1 5 3 3 3 3 3 3 3 2 3 3 3 3 3 5 3 3 3 3 1 3 3 3 3 3 3 3 3 3 6 3
#> [39] 3 6 1 3 3 1 3 3 3 3 3 3 3 1 3 3 3 3 3 1 3 3 1 1 1 3 3 3 1 3 3 1 1 3 4 3 3 1
#> [77] 3 3 3 6 3 3 1 3 5 6 1And which rows each group contains with group_rows():
by_sex_gender %>% group_rows()
#> <list_of<integer>[6]>
#> [[1]]
#> [1] 5 7 27 41 44 52 58 61 62 63 67 70 71 76 83 87
#>
#> [[2]]
#> [1] 16
#>
#> [[3]]
#> [1] 1 4 6 9 10 11 12 13 14 15 17 18 19 20 21 23 24 25 26 28 29 30 31 32 33
#> [26] 34 35 36 38 39 42 43 45 46 47 48 49 50 51 53 54 55 56 57 59 60 64 65 66 68
#> [51] 69 72 74 75 77 78 79 81 82 84
#>
#> [[4]]
#> [1] 73
#>
#> [[5]]
#> [1] 2 3 8 22 85
#>
#> [[6]]
#> [1] 37 40 80 86group_size() gives the size of each group:
by_sex_gender %>% group_size()
#> [1] 16 1 60 1 5 4n_groups() gives the total number of groups:
by_sex_gender %>% n_groups()
#> [1] 6Use group_vars() if you just want the names of the grouping variables:
by_species %>% group_vars()
#> [1] "species"
by_sex_gender %>% group_vars()
#> [1] "sex" "gender"6.2.2 “current” group information
Inside dplyr verbs, you can access various properties of the “current” group using a family of functions with the cur_ prefix. These functions are typically needed for everyday usage of dplyr, but can be useful because they allow you to free from some of the typical constraints of dplyr verbs.
df <- tibble(
g1 = c('a', 'a', 'a', 'b', 'b', 'b'),
g2 = c('c', 'c', 'd', 'e', 'e', 'e'),
x = 1:6,
y = 2:7
)
gf <- df %>% group_by(g1, g2)
gf
#> # A tibble: 6 x 4
#> # Groups: g1, g2 [3]
#> g1 g2 x y
#> <chr> <chr> <int> <int>
#> 1 a c 1 2
#> 2 a c 2 3
#> 3 a d 3 4
#> 4 b e 4 5
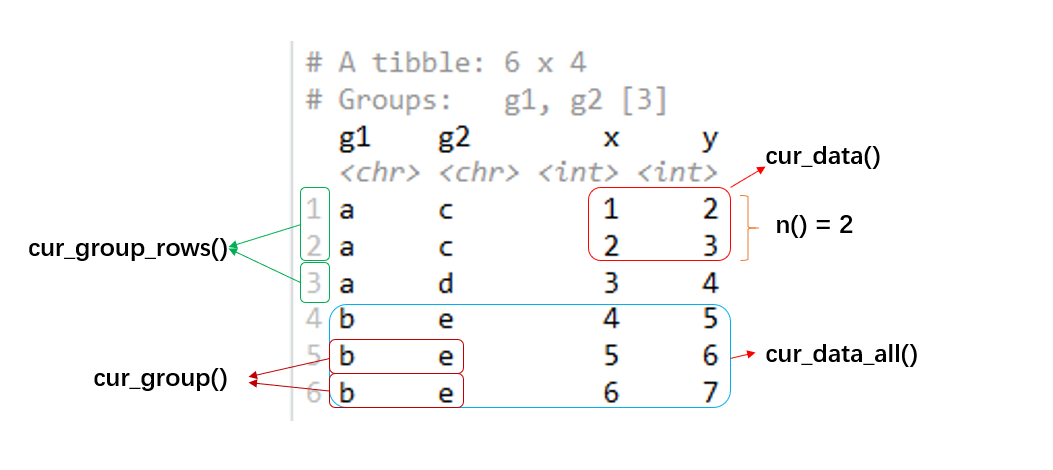
#> # … with 2 more rowsn()gives the current group size.gf %>% summarise(n = n()) #> `summarise()` has grouped output by 'g1'. You can override using the `.groups` argument. #> # A tibble: 3 x 3 #> # Groups: g1 [2] #> g1 g2 n #> <chr> <chr> <int> #> 1 a c 2 #> 2 a d 1 #> 3 b e 3cur_data()gives the current data for the current group (excluding grouping variables).gf %>% summarise(data = list(cur_data())) #> `summarise()` has grouped output by 'g1'. You can override using the `.groups` argument. #> # A tibble: 3 x 3 #> # Groups: g1 [2] #> g1 g2 data #> <chr> <chr> <list> #> 1 a c <tibble[,2] [2 × 2]> #> 2 a d <tibble[,2] [1 × 2]> #> 3 b e <tibble[,2] [3 × 2]>cur_data_all()gives the current data for the current group (including grouping variables).gf %>% summarise(data = list(cur_data_all())) #> `summarise()` has grouped output by 'g1'. You can override using the `.groups` argument. #> # A tibble: 3 x 3 #> # Groups: g1 [2] #> g1 g2 data #> <chr> <chr> <list> #> 1 a c <tibble[,4] [2 × 4]> #> 2 a d <tibble[,4] [1 × 4]> #> 3 b e <tibble[,4] [3 × 4]>cur_group()gives the group keys, a tibble with one row and one column for each grouping variable.gf %>% summarise(data = list(cur_group())) #> `summarise()` has grouped output by 'g1'. You can override using the `.groups` argument. #> # A tibble: 3 x 3 #> # Groups: g1 [2] #> g1 g2 data #> <chr> <chr> <list> #> 1 a c <tibble[,2] [1 × 2]> #> 2 a d <tibble[,2] [1 × 2]> #> 3 b e <tibble[,2] [1 × 2]>cur_group_id()gives a unique numeric identifier for the current group.gf %>% mutate(id = cur_group_id()) #> # A tibble: 6 x 5 #> # Groups: g1, g2 [3] #> g1 g2 x y id #> <chr> <chr> <int> <int> <int> #> 1 a c 1 2 1 #> 2 a c 2 3 1 #> 3 a d 3 4 2 #> 4 b e 4 5 3 #> # … with 2 more rowscur_group_rows()gives the row indices for the current group.gf %>% summarise(data = list(cur_group_rows())) #> `summarise()` has grouped output by 'g1'. You can override using the `.groups` argument. #> # A tibble: 3 x 3 #> # Groups: g1 [2] #> g1 g2 data #> <chr> <chr> <list> #> 1 a c <int [2]> #> 2 a d <int [1]> #> 3 b e <int [3]>
You can easily distinguish these functions in the following picture:
6.3 How group_by() works
group_by() create a new class of objects, grouped_df, who is treated differently with tbl_df, in most dplyr verbs.
df <- tibble(
g1 = c('b', 'b', 'b', 'a', 'a', 'a'),
g2 = c('c', 'c', 'd', 'e', 'e', 'e'),
x = 1:6
)
gf <- df %>% group_by(g1, g2)
df %>% class()
#> [1] "tbl_df" "tbl" "data.frame"
gf %>% class()
#> [1] "grouped_df" "tbl_df" "tbl" "data.frame"But group_by() never change the value of datasets (even the order of rows):
df
#> # A tibble: 6 x 3
#> g1 g2 x
#> <chr> <chr> <int>
#> 1 b c 1
#> 2 b c 2
#> 3 b d 3
#> 4 a e 4
#> # … with 2 more rows
gf
#> # A tibble: 6 x 3
#> # Groups: g1, g2 [3]
#> g1 g2 x
#> <chr> <chr> <int>
#> 1 b c 1
#> 2 b c 2
#> 3 b d 3
#> 4 a e 4
#> # … with 2 more rows
df == gf
#> g1 g2 x
#> [1,] TRUE TRUE TRUE
#> [2,] TRUE TRUE TRUE
#> [3,] TRUE TRUE TRUE
#> [4,] TRUE TRUE TRUE
#> [5,] TRUE TRUE TRUE
#> [6,] TRUE TRUE TRUEHere we can easily notice that dplyr verbs have different behavior for df and gf:
df %>% mutate(sum = sum(x))
#> # A tibble: 6 x 4
#> g1 g2 x sum
#> <chr> <chr> <int> <int>
#> 1 b c 1 21
#> 2 b c 2 21
#> 3 b d 3 21
#> 4 a e 4 21
#> # … with 2 more rows
gf %>% mutate(sum = sum(x))
#> # A tibble: 6 x 4
#> # Groups: g1, g2 [3]
#> g1 g2 x sum
#> <chr> <chr> <int> <int>
#> 1 b c 1 3
#> 2 b c 2 3
#> 3 b d 3 3
#> 4 a e 4 15
#> # … with 2 more rowsBut in other places we should be cautious when we use a dataset output from a wrapped function, or write a function who receive an user-supplied data. For not knowing whether a grouped_df you get (or receive).
7 Row-wise operations
dplyr, and R in general, are particularly well suited to performing operations over columns, and performing operations over rows is much harder.
Let’s start from a little problem. We create a tibble df and want to calculate the sum of w x y z in each row:
df <- tibble(id = 1:6, w = 10:15, x = 20:25, y = 30:35, z = 40:45)
df %>% mutate(total = sum(c(w, x, y, z)))
#> # A tibble: 6 x 6
#> id w x y z total
#> <int> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40 660
#> 2 2 11 21 31 41 660
#> 3 3 12 22 32 42 660
#> 4 4 13 23 33 43 660
#> # … with 2 more rowsIt doesn’t work out as expected, total calculate the sum of all value from w x y z (not in each row). This is because that sum() just return a single value calculated from the input:
sum(c(df$w, df$x, df$y, df$z))
#> [1] 660then tibble treat the input as total = 660 and automatically recycle it.
The same problem will occur if you want to do row-wise aggregates with mean(), min(), max() and other functions who are not vectorised function. In order to solve this problem, you can use dplyr’s approach centred around the row-wise data frame created by rowwise().
7.1 rowwise()
Row-wise operations require a special type of grouping where each group consists of a single row. You create this with rowwise():
df %>% rowwise()
#> # A tibble: 6 x 5
#> # Rowwise:
#> id w x y z
#> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40
#> 2 2 11 21 31 41
#> 3 3 12 22 32 42
#> 4 4 13 23 33 43
#> # … with 2 more rowsLike group_by(), rowwise() doesn’t really do anything itself; it just changes how the other verbs work. For example, compare the results of mutate() in the following code:
df %>% mutate(total = sum(c(w, x, y, z)))
#> # A tibble: 6 x 6
#> id w x y z total
#> <int> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40 660
#> 2 2 11 21 31 41 660
#> 3 3 12 22 32 42 660
#> 4 4 13 23 33 43 660
#> # … with 2 more rows
df %>% rowwise() %>% mutate(total = sum(c(w, x, y, z)))
#> # A tibble: 6 x 6
#> # Rowwise:
#> id w x y z total
#> <int> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40 100
#> 2 2 11 21 31 41 104
#> 3 3 12 22 32 42 108
#> 4 4 13 23 33 43 112
#> # … with 2 more rowsIf you use mutate() with a regular data frame, it computes the sum of w, x, y, and z across all rows. If you apply it to a row-wise data frame, it computes the sum for each row.
You can optionally supply “identifier” variables in your call to rowwise(). These variables are preserved when you call summarise(), so they behave somewhat similarly to the grouping variables passed to group_by():
df %>%
rowwise() %>%
summarise(total = sum(c(w, x, y, z)))
#> `summarise()` has ungrouped output. You can override using the `.groups` argument.
#> # A tibble: 6 x 1
#> total
#> <int>
#> 1 100
#> 2 104
#> 3 108
#> 4 112
#> # … with 2 more rows
df %>%
rowwise(id) %>%
summarise(total = sum(c(w, x, y, z)))
#> `summarise()` has grouped output by 'id'. You can override using the `.groups` argument.
#> # A tibble: 6 x 2
#> # Groups: id [6]
#> id total
#> <int> <int>
#> 1 1 100
#> 2 2 104
#> 3 3 108
#> 4 4 112
#> # … with 2 more rowsrowwise() is just a special form of grouping, so if you want to remove it from a data frame, just call ungroup().
You can use c_across() which uses tidy selection syntax so you can to succinctly select many variables:
df %>%
rowwise() %>%
mutate(total = sum(c_across(w:z)))
#> # A tibble: 6 x 6
#> # Rowwise:
#> id w x y z total
#> <int> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40 100
#> 2 2 11 21 31 41 104
#> 3 3 12 22 32 42 108
#> 4 4 13 23 33 43 112
#> # … with 2 more rows7.2 Other solution
The rowwise() approach will work for any summary function. But if you need greater speed, it’s worth looking for a built-in row-wise variant of your summary function. These are more efficient because they operate on the data frame as whole; they don’t split it into rows, compute the summary, and then join the results back together again.
df %>% mutate(total = rowSums(across(w:z)))
#> # A tibble: 6 x 6
#> id w x y z total
#> <int> <int> <int> <int> <int> <dbl>
#> 1 1 10 20 30 40 100
#> 2 2 11 21 31 41 104
#> 3 3 12 22 32 42 108
#> 4 4 13 23 33 43 112
#> # … with 2 more rows
df %>% mutate(mean = rowMeans(across(w:z)))
#> # A tibble: 6 x 6
#> id w x y z mean
#> <int> <int> <int> <int> <int> <dbl>
#> 1 1 10 20 30 40 25
#> 2 2 11 21 31 41 26
#> 3 3 12 22 32 42 27
#> 4 4 13 23 33 43 28
#> # … with 2 more rowsNote: we don’t use rowwise() and use across() in rowMeans() and rowSums() here because across() returns a tibble and rowMeans() and rowSums() take a multi-row data frame as input.
Or you can use vectorised function, such as pmin(), pmax(). Similarly, they’ll be much faster than rowwise().
df %>% mutate(
min = pmin(w, x, y, z),
max = pmax(w, x, y, z)
)
#> # A tibble: 6 x 7
#> id w x y z min max
#> <int> <int> <int> <int> <int> <int> <int>
#> 1 1 10 20 30 40 10 40
#> 2 2 11 21 31 41 11 41
#> 3 3 12 22 32 42 12 42
#> 4 4 13 23 33 43 13 43
#> # … with 2 more rows7.3 Difference between c_across(), across(), if_any() and if_all()
You can view the output structure of c_across(), across(), if_any() and if_all() by wrapping with list():
df %>% rowwise() %>% mutate(new = list(c_across(w:z)))
#> # A tibble: 6 x 6
#> # Rowwise:
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <int [4]>
#> 2 2 11 21 31 41 <int [4]>
#> 3 3 12 22 32 42 <int [4]>
#> 4 4 13 23 33 43 <int [4]>
#> # … with 2 more rows
df %>% rowwise() %>% mutate(new = list(across(w:z)))
#> # A tibble: 6 x 6
#> # Rowwise:
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <tibble[,4] [1 × 4]>
#> 2 2 11 21 31 41 <tibble[,4] [1 × 4]>
#> 3 3 12 22 32 42 <tibble[,4] [1 × 4]>
#> 4 4 13 23 33 43 <tibble[,4] [1 × 4]>
#> # … with 2 more rows
df %>% rowwise() %>% mutate(new = list(if_any(w:z, ~ .x > 25)))
#> # A tibble: 6 x 6
#> # Rowwise:
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <lgl [1]>
#> 2 2 11 21 31 41 <lgl [1]>
#> 3 3 12 22 32 42 <lgl [1]>
#> 4 4 13 23 33 43 <lgl [1]>
#> # … with 2 more rows
df %>% mutate(new = list(c_across(w:z)))
#> # A tibble: 6 x 6
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <int [24]>
#> 2 2 11 21 31 41 <int [24]>
#> 3 3 12 22 32 42 <int [24]>
#> 4 4 13 23 33 43 <int [24]>
#> # … with 2 more rows
df %>% mutate(new = list(across(w:z)))
#> # A tibble: 6 x 6
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <tibble[,4] [6 × 4]>
#> 2 2 11 21 31 41 <tibble[,4] [6 × 4]>
#> 3 3 12 22 32 42 <tibble[,4] [6 × 4]>
#> 4 4 13 23 33 43 <tibble[,4] [6 × 4]>
#> # … with 2 more rows
df %>% mutate(new = list(if_any(w:z, ~ .x > 25)))
#> # A tibble: 6 x 6
#> id w x y z new
#> <int> <int> <int> <int> <int> <list>
#> 1 1 10 20 30 40 <lgl [6]>
#> 2 2 11 21 31 41 <lgl [6]>
#> 3 3 12 22 32 42 <lgl [6]>
#> 4 4 13 23 33 43 <lgl [6]>
#> # … with 2 more rowsc_across() returns a vector, across() returns a tibble, if_any() and if_all() return a logical vector. We can also find that c_across() doesn’t work well without rowwise(). Use them cautiously depending on other function’s needed input.
8 Vector functions
8.1 Vectorised if: if_else() case_when()
if_else(condition, true, false, missing = NULL)
case_when(...)
x <- c(-5:5, NA)
if_else(x < 0, true = "negative", false = "positive", missing = "missing")
#> [1] "negative" "negative" "negative" "negative" "negative" "positive"
#> [7] "positive" "positive" "positive" "positive" "positive" "missing"
x <- 1:50
case_when(
x %% 35 == 0 ~ "fizz buzz",
x %% 5 == 0 ~ "fizz",
x %% 7 == 0 ~ "buzz",
TRUE ~ as.character(x)
)
#> [1] "1" "2" "3" "4" "fizz" "6"
#> [7] "buzz" "8" "9" "fizz" "11" "12"
#> [13] "13" "buzz" "fizz" "16" "17" "18"
#> [19] "19" "fizz" "buzz" "22" "23" "24"
#> [25] "fizz" "26" "27" "buzz" "29" "fizz"
#> [31] "31" "32" "33" "34" "fizz buzz" "36"
#> [37] "37" "38" "39" "fizz" "41" "buzz"
#> [43] "43" "44" "fizz" "46" "47" "48"
#> [49] "buzz" "fizz"If none of the cases match in case_when() (or do not clarify missing in if_else()), NA is used as return value:
x <- c(-1, 1, NA)
if_else(x < 0, 'negative', 'positive')
#> [1] "negative" "positive" NA
case_when(
x < 0 ~ 'negative',
x >= 0 ~ 'positive'
)
#> [1] "negative" "positive" NAUse %>% with if_else() and case_when() (wrapped them with {}):
x <- c(-1, 1, NA)
x %>% {
if_else(. < 0, "negative", "positive", "missing")
}
#> [1] "negative" "positive" "missing"
x %>% {
case_when(
. < 0 ~ "negative",
. >= 0 ~ "positive",
TRUE ~ "missing"
)
}
#> [1] "negative" "positive" "missing"Note that if_else() and case_when() are both strict, if_else() checks that true and false are the same type, in case_when(), all RHSs must evaluate to the same type of vector. Inconsistent types will throw an error. This applies also to NA values used in RHS (or true false): NA is logical, use typed values like NA_real_, NA_complex_, NA_character_, NA_integer_ as appropriate.
x <- c(-1, 1)
case_when(
x >= 0 ~ "positive",
TRUE ~ NA
)
#> Error: must be a character vector, not a logical vector.
case_when(
x >= 0 ~ "positive",
TRUE ~ NA_character_
)
#> [1] NA "positive"if_else() and case_when() evaluates all expressions first, and then constructs its result by extracting the selected parts.
x <- -2:2 %>% as.double()
if_else(x >= 0, sqrt(x), x)
#> Warning in sqrt(x): NaNs produced
#> [1] -2.000000 -1.000000 0.000000 1.000000 1.414214
case_when(
x >= 0 ~ sqrt(x),
TRUE ~ x
)
#> Warning in sqrt(x): NaNs produced
#> [1] -2.000000 -1.000000 0.000000 1.000000 1.414214Sometimes it’s hard to generate multiple variables in complex condition statement via if_else() and case_when(), you can turn to use libr::datastep():
df <- tibble(x = 1:3, y = 4:6)
df %>% libr::datastep({
if (x <= 2 & y == 4) {
var1 <- 'a'
var_temp <- 1
} else {
var1 <- 'b'
var_temp <- 0
}
if (var_temp == 1 | x == 3) {
var2 <- 'c'
}
rm(var_temp)
})
#> # A tibble: 3 x 4
#> x y var2 var1
#> <int> <int> <chr> <chr>
#> 1 1 4 c a
#> 2 2 5 <NA> b
#> 3 3 6 c b8.2 Cumulativate functions: cumall() cumany() cummean()
Cumulative aggregates: cumsum(), cumprod(), cummin(), cummax() (from base R), and cumall(), cumany(), and cummean() (from dplyr).
cumsum(1:10)
#> [1] 1 3 6 10 15 21 28 36 45 55
cumprod(1:5)
#> [1] 1 2 6 24 120
cummean(10:1)
#> [1] 10.0 9.5 9.0 8.5 8.0 7.5 7.0 6.5 6.0 5.5
cummin(c(3, 2, 1, 2, 1, 0, 4, 3, 2))
#> [1] 3 2 1 1 1 0 0 0 0
cummax(c(3, 2, 1, 2, 1, 0, 4, 3, 2))
#> [1] 3 3 3 3 3 3 4 4 4
cumall(c(T, T, F, T, F))
#> [1] TRUE TRUE TRUE TRUE TRUE
cumany(c(F, F, T, F, T))
#> [1] TRUE TRUE TRUE TRUE TRUECumulative logical functions cumall() and cumany() are particularly useful in conjunction with filter():
cumall(x): all cases until the firstFALSE.cumall(!x): all cases until the firstTRUE.cumany(x): all cases after the firstTRUE.cumany(!x): all cases after the firstFALSE.
df <- data.frame(
date = as.Date("2020-01-01") + 0:6,
balance = c(100, 50, 25, -25, -50, 30, 120)
)
df
#> date balance
#> 1 2020-01-01 100
#> 2 2020-01-02 50
#> 3 2020-01-03 25
#> 4 2020-01-04 -25
#> 5 2020-01-05 -50
#> 6 2020-01-06 30
#> 7 2020-01-07 120
# all rows after first overdraft
df %>% filter(cumany(balance < 0))
#> date balance
#> 1 2020-01-04 -25
#> 2 2020-01-05 -50
#> 3 2020-01-06 30
#> 4 2020-01-07 120
# all rows until first overdraft
df %>% filter(cumall(!(balance < 0)))
#> date balance
#> 1 2020-01-01 100
#> 2 2020-01-02 50
#> 3 2020-01-03 258.3 Windowed rank functions: row_number() ntile() min_rank() dense_rank() percent_rank() cume_dist()
row_number(): equivalent torank(ties.method = "first")min_rank(): equivalent torank(ties.method = "min")dense_rank(): likemin_rank(), but with no gaps between rankspercent_rank(): a number between 0 and 1 computed by rescalingmin_rank()to[0, 1]cume_dist(): a cumulative distribution function. Proportion of all values less than or equal to the current rank.ntile(): a rough rank, which breaks the input vector intonbuckets. The size of the buckets may differ by up to one, larger buckets have lower rank.
x <- c(5, 1, 3, 2, 2)
row_number(x)
#> [1] 5 1 4 2 3
min_rank(x)
#> [1] 5 1 4 2 2
dense_rank(x)
#> [1] 4 1 3 2 2
percent_rank(x)
#> [1] 1.00 0.00 0.75 0.25 0.25
cume_dist(x)
#> [1] 1.0 0.2 0.8 0.6 0.6
ntile(x, 2)
#> [1] 2 1 2 1 1
ntile(1:8, 3)
#> [1] 1 1 1 2 2 2 3 3All ranking functions rank from lowest to highest so that small input values get small ranks. Use desc() to rank from highest to lowest.
x <- c(5, 1, 3, 2, 2)
row_number(desc(x))
#> [1] 1 5 2 3 4
min_rank(desc(x))
#> [1] 1 5 2 3 3
dense_rank(desc(x))
#> [1] 1 4 2 3 3
percent_rank(desc(x))
#> [1] 0.00 1.00 0.25 0.50 0.50
cume_dist(desc(x))
#> [1] 0.2 1.0 0.4 0.8 0.8
ntile(desc(x), 2)
#> [1] 1 2 1 1 2row_number() can be used with single table verbs without specifying x to denote the row number, and it would be affected by group_by():
starwars %>%
select(name, species) %>%
mutate(n = row_number())
#> # A tibble: 87 x 3
#> name species n
#> <chr> <chr> <int>
#> 1 Luke Skywalker Human 1
#> 2 C-3PO Droid 2
#> 3 R2-D2 Droid 3
#> 4 Darth Vader Human 4
#> # … with 83 more rows
starwars %>%
select(name, species) %>%
group_by(species) %>%
mutate(n = row_number())
#> # A tibble: 87 x 3
#> # Groups: species [38]
#> name species n
#> <chr> <chr> <int>
#> 1 Luke Skywalker Human 1
#> 2 C-3PO Droid 1
#> 3 R2-D2 Droid 2
#> 4 Darth Vader Human 2
#> # … with 83 more rows8.4 Extract the first, last or nth value from a vector: nth() first() last()
first(x) last(x) nth(x, n)
x <- 1:10
first(x)
#> [1] 1
last(x)
#> [1] 10
nth(x, 1)
#> [1] 1
nth(x, 5)
#> [1] 5
nth(x, -2)
#> [1] 9
nth(x, 11)
#> [1] NA8.5 Compute lagged or leading values: lag() lead()
lag(x, n = 1L, default = NA)
lead(x, n = 1L, default = NA)
lag(1:5)
#> [1] NA 1 2 3 4
lead(1:5)
#> [1] 2 3 4 5 NA
lag(1:5, n = 1)
#> [1] NA 1 2 3 4
lag(1:5, n = 2)
#> [1] NA NA 1 2 3
lead(1:5, n = 1)
#> [1] 2 3 4 5 NA
lead(1:5, n = 2)
#> [1] 3 4 5 NA NA
lag(1:5, default = 0)
#> [1] 0 1 2 3 4
lead(1:5, default = 6)
#> [1] 2 3 4 5 68.6 Find first non-missing element: coalesce()
y <- c(1, 2, NA, NA, 5, NA)
z <- c(NA, NA, 3, 4, 5, NA)
coalesce(y, z)
#> [1] 1 2 3 4 5 NAYou can use a single value to replace all missing values:
x <- sample(c(1:5, NA, NA, NA))
x
#> [1] NA NA 2 NA 5 1 3 4
coalesce(x, 0L)
#> [1] 0 0 2 0 5 1 3 48.7 Convert values to NA: na_if()
na_if(x, y)
x: Vector to modifyy: Value to replace withNA
na_if(1:5, 2)
#> [1] 1 NA 3 4 5
na_if(1:5, 5:1)
#> [1] 1 2 NA 4 58.8 Compare two numeric vectors: near()
near(x, y, tol = .Machine$double.eps^0.5)
sqrt(2) ^ 2 == 2
#> [1] FALSE
near(sqrt(2) ^ 2, 2)
#> [1] TRUE8.9 Shortcut of left <= x & x <= right: between()
between(x, left, right)
between(1:12, 7, 9)
#> [1] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE FALSE FALSE FALSE8.10 Efficiently count the number of unique values in a set of vector: n_distinct()
n_distinct(..., na.rm = FALSE)
x <- sample(1:10, 1e7, rep = TRUE)
n_distinct(x)
#> [1] 10
length(unique(x)) %>% system.time()
#> user system elapsed
#> 0.212 0.036 0.248
n_distinct(x) %>% system.time()
#> user system elapsed
#> 0.077 0.023 0.101