sqldf
Description
library(sqldf)
#> Loading required package: gsubfn
#> Loading required package: proto
#> Warning in fun(libname, pkgname): couldn't connect to display ":0"
#> Loading required package: RSQLiteThe sqldf() function is typically passed a single argument which is an SQL select statement where the table names are ordinary R data frame names. sqldf() transparently sets up a database, imports the data frames into that database, performs the SQL select or other statement and returns the result using a heuristic to determine which class to assign to each column of the returned data frame. The sqldf() or read.csv.sql() functions can also be used to read filtered files into R even if the original files are larger than R itself can handle.
‘RSQLite’, ‘RH2’, ‘RMySQL’ and ‘RPostgreSQL’ backends are supported.
sqldf Usage
Read a file by read.csv.sql()
Read a file into R filtering it with an sql statement. Only the filtered portion is processed by R so that files larger than R can otherwise handle can be accommodated. read_sql.csv
df <- read.csv.sql(
'./data/read_sql.csv',
sql = "select * from file",
header = TRUE,
sep = ","
)
head(df)
#> SubjectName status Ordinal Display.f subjid color
#> 1 066001-001 1 1 SCR 5 green\r
#> 2 066001-001 1 2 C1D1 5 green\r
#> 3 066001-001 1 3 C1D8 5 green\r
#> 4 066001-001 1 4 C1D15 5 green\r
#> 5 066001-001 1 5 C2D1 5 green\r
#> 6 066001-001 1 53 EOT 5 green\rcreate a data frame
df <- data.frame(a = 1:5, b = letters[1:5])
sqldf("select * from df")
#> a b
#> 1 1 a
#> 2 2 b
#> 3 3 c
#> 4 4 d
#> 5 5 e
sqldf("select avg(a) mean, variance(a) var from df")
#> mean var
#> 1 3 2.5DF <- data.frame(
index=1:12,
date=rep(c(Sys.Date() - 1, Sys.Date()), 6),
group=c("A", "B", "C"),
value=round(rnorm(12), 2)
)
sqldf("select * from DF")
#> index date group value
#> 1 1 2021-10-24 A 0.78
#> 2 2 2021-10-25 B 0.33
#> 3 3 2021-10-24 C 0.13
#> 4 4 2021-10-25 A 0.08
#> 5 5 2021-10-24 B -0.41
#> 6 6 2021-10-25 C -1.78
#> 7 7 2021-10-24 A 0.27
#> 8 8 2021-10-25 B -1.60
#> 9 9 2021-10-24 C 1.37
#> 10 10 2021-10-25 A 1.57
#> 11 11 2021-10-24 B -0.10
#> 12 12 2021-10-25 C -0.27select var from dataset
# select all vars from iris dataset
df <- sqldf('select * from iris')
head(df)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3.0 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5.0 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa
# select "Sepal.Length", Species from iris dataset
df <- sqldf('select Species, "Sepal.Length" from iris ')
head(df)
#> Species Sepal.Length
#> 1 setosa 5.1
#> 2 setosa 4.9
#> 3 setosa 4.7
#> 4 setosa 4.6
#> 5 setosa 5.0
#> 6 setosa 5.4What will happen when running the below code?
sqldf('select Species, Sepal.Length from iris')head in basic R vs limit in sql
a1r <- head(warpbreaks, 6)
a1r
#> breaks wool tension
#> 1 26 A L
#> 2 30 A L
#> 3 54 A L
#> 4 25 A L
#> 5 70 A L
#> 6 52 A L
a1s <- sqldf("select * from warpbreaks limit 6")
a1s
#> breaks wool tension
#> 1 26 A L
#> 2 30 A L
#> 3 54 A L
#> 4 25 A L
#> 5 70 A L
#> 6 52 A L
identical(a1r, a1s)
#> [1] TRUEfilter record by where or having stament
df1 <- sqldf('select * from iris where "Sepal.Width" > 3')
head(df1)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.7 3.2 1.3 0.2 setosa
#> 3 4.6 3.1 1.5 0.2 setosa
#> 4 5.0 3.6 1.4 0.2 setosa
#> 5 5.4 3.9 1.7 0.4 setosa
#> 6 4.6 3.4 1.4 0.3 setosa
df2 <- sqldf('select * from iris group by Species having "Sepal.Width" > 3')
df2
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 7.0 3.2 4.7 1.4 versicolor
#> 3 6.3 3.3 6.0 2.5 virginicacreate new var
df2 <- sqldf('
select Species, "Sepal.Length", avg("Sepal.Length") `col1`
from iris
group by Species
order by Species
')
df2
#> Species Sepal.Length col1
#> 1 setosa 5.1 5.006
#> 2 versicolor 7.0 5.936
#> 3 virginica 6.3 6.588
df21 <- sqldf("
select Species, 'Sepal.Length', 'new' as col1
from iris
group by Species
order by Species
")
df21
#> Species 'Sepal.Length' col1
#> 1 setosa Sepal.Length new
#> 2 versicolor Sepal.Length new
#> 3 virginica Sepal.Length newcreate new var by case when
df3 <- sqldf('
select Species, "Sepal.Length"
from iris
group by Species
order by Species
')
df3
#> Species Sepal.Length
#> 1 setosa 5.1
#> 2 versicolor 7.0
#> 3 virginica 6.3
df4 <- sqldf("
select Species, 'Sepal.Length',
CASE
WHEN Species = 'setosa' THEN 'specy1'
ELSE 'other'
END as setosa,
case
when Species = 'setosa' then 1
else 0
end as setosan
from iris
")
head(df4)
#> Species 'Sepal.Length' setosa setosan
#> 1 setosa Sepal.Length specy1 1
#> 2 setosa Sepal.Length specy1 1
#> 3 setosa Sepal.Length specy1 1
#> 4 setosa Sepal.Length specy1 1
#> 5 setosa Sepal.Length specy1 1
#> 6 setosa Sepal.Length specy1 1group by, order by
df <- sqldf('select * from iris')
df0 <- sqldf('select * from df group by Species')
df0
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 7.0 3.2 4.7 1.4 versicolor
#> 3 6.3 3.3 6.0 2.5 virginica
df01 <- sqldf('select * from df group by "Sepal.Length"')
head(df01)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.3 3.0 1.1 0.1 setosa
#> 2 4.4 2.9 1.4 0.2 setosa
#> 3 4.5 2.3 1.3 0.3 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 4.7 3.2 1.3 0.2 setosa
#> 6 4.8 3.4 1.6 0.2 setosa
df1 <- sqldf('select * from df group by Species order by "Sepal.Length" desc, Species')
df1
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 7.0 3.2 4.7 1.4 versicolor
#> 2 6.3 3.3 6.0 2.5 virginica
#> 3 5.1 3.5 1.4 0.2 setosadistinct : eliminate duplicate records
sqldf("select DISTINCT Species from iris")
#> Species
#> 1 setosa
#> 2 versicolor
#> 3 virginicaexcept
one <- head(iris)
two <- iris
nrow(one)
#> [1] 6
nrow(two)
#> [1] 150
# if a and not b
EXCEPT <- sqldf("select * from one EXCEPT select * from two")
EXCEPT
#> [1] Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <0 rows> (or 0-length row.names)
EXCEPT2 <- sqldf("select * from two EXCEPT select * from one")
nrow(EXCEPT2)
#> [1] 143
head(EXCEPT2)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.3 3.0 1.1 0.1 setosa
#> 2 4.4 2.9 1.4 0.2 setosa
#> 3 4.4 3.0 1.3 0.2 setosa
#> 4 4.4 3.2 1.3 0.2 setosa
#> 5 4.5 2.3 1.3 0.3 setosa
#> 6 4.6 3.2 1.4 0.2 setosaintersect
INTERSECT <- sqldf("select * from one INTERSECT select * from two")
INTERSECT
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 4.6 3.1 1.5 0.2 setosa
#> 2 4.7 3.2 1.3 0.2 setosa
#> 3 4.9 3.0 1.4 0.2 setosa
#> 4 5.0 3.6 1.4 0.2 setosa
#> 5 5.1 3.5 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosaSQL Joins
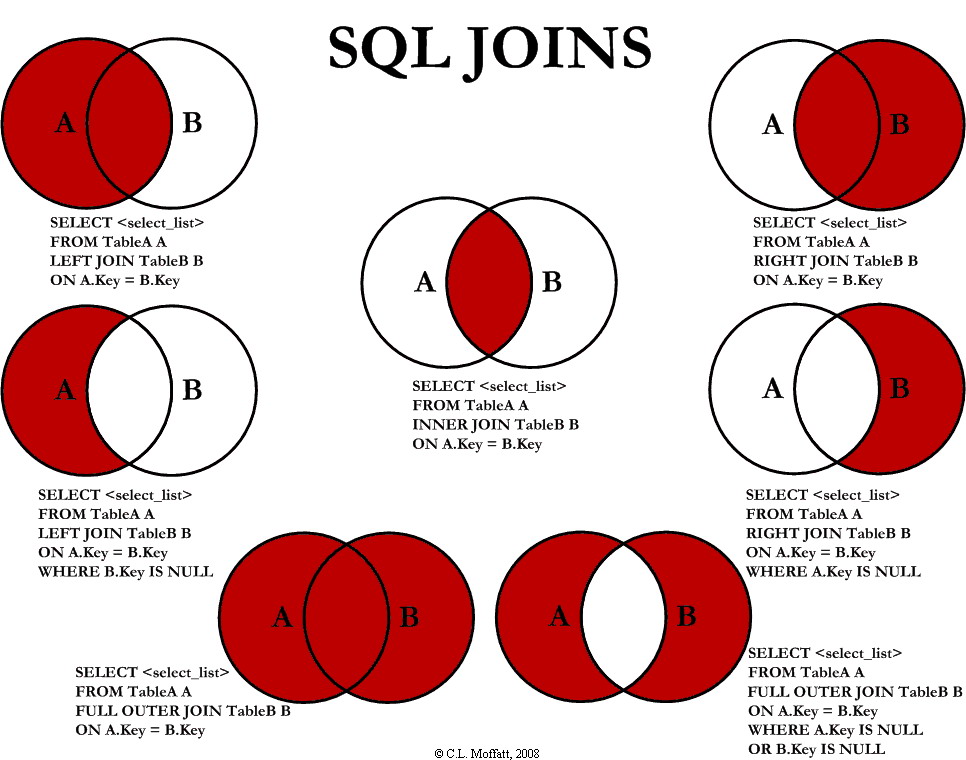
inner join
df1 <- head(iris)
df2 <- iris
# 筛选相同id,all = F为只连接匹配到的,all = T为没有匹配到的赋值NA
inner1 <- merge(df1, df2, by = "Species", all = F)
head(inner1)
#> Species Sepal.Length.x Sepal.Width.x Petal.Length.x Petal.Width.x
#> 1 setosa 5.1 3.5 1.4 0.2
#> 2 setosa 5.1 3.5 1.4 0.2
#> 3 setosa 5.1 3.5 1.4 0.2
#> 4 setosa 5.1 3.5 1.4 0.2
#> 5 setosa 5.1 3.5 1.4 0.2
#> 6 setosa 5.1 3.5 1.4 0.2
#> Sepal.Length.y Sepal.Width.y Petal.Length.y Petal.Width.y
#> 1 5.4 3.9 1.7 0.4
#> 2 5.0 3.6 1.4 0.2
#> 3 4.9 3.1 1.5 0.1
#> 4 4.6 3.4 1.4 0.3
#> 5 5.0 3.4 1.5 0.2
#> 6 4.4 2.9 1.4 0.2
# 内连接
inner3 <- sqldf("select * from df1 as a inner join df2 as b on a.Species=b.Species")
head(inner3)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species Sepal.Length
#> 1 5.1 3.5 1.4 0.2 setosa 4.3
#> 2 5.1 3.5 1.4 0.2 setosa 4.4
#> 3 5.1 3.5 1.4 0.2 setosa 4.4
#> 4 5.1 3.5 1.4 0.2 setosa 4.4
#> 5 5.1 3.5 1.4 0.2 setosa 4.5
#> 6 5.1 3.5 1.4 0.2 setosa 4.6
#> Sepal.Width Petal.Length Petal.Width Species
#> 1 3.0 1.1 0.1 setosa
#> 2 2.9 1.4 0.2 setosa
#> 3 3.0 1.3 0.2 setosa
#> 4 3.2 1.3 0.2 setosa
#> 5 2.3 1.3 0.3 setosa
#> 6 3.1 1.5 0.2 setosa
# 笛卡尔积
inner4 <- sqldf("select * from df1 as a,df2 as b where a.Species=b.Species")
head(inner4)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species Sepal.Length
#> 1 5.1 3.5 1.4 0.2 setosa 4.3
#> 2 5.1 3.5 1.4 0.2 setosa 4.4
#> 3 5.1 3.5 1.4 0.2 setosa 4.4
#> 4 5.1 3.5 1.4 0.2 setosa 4.4
#> 5 5.1 3.5 1.4 0.2 setosa 4.5
#> 6 5.1 3.5 1.4 0.2 setosa 4.6
#> Sepal.Width Petal.Length Petal.Width Species
#> 1 3.0 1.1 0.1 setosa
#> 2 2.9 1.4 0.2 setosa
#> 3 3.0 1.3 0.2 setosa
#> 4 3.2 1.3 0.2 setosa
#> 5 2.3 1.3 0.3 setosa
#> 6 3.1 1.5 0.2 setosa
identical(inner3, inner4)
#> [1] TRUEleft join
left1 <- merge(df1, df2, by = "Species", all.x = TRUE)
head(left1)
#> Species Sepal.Length.x Sepal.Width.x Petal.Length.x Petal.Width.x
#> 1 setosa 5.1 3.5 1.4 0.2
#> 2 setosa 5.1 3.5 1.4 0.2
#> 3 setosa 5.1 3.5 1.4 0.2
#> 4 setosa 5.1 3.5 1.4 0.2
#> 5 setosa 5.1 3.5 1.4 0.2
#> 6 setosa 5.1 3.5 1.4 0.2
#> Sepal.Length.y Sepal.Width.y Petal.Length.y Petal.Width.y
#> 1 5.4 3.9 1.7 0.4
#> 2 5.0 3.6 1.4 0.2
#> 3 4.9 3.1 1.5 0.1
#> 4 4.6 3.4 1.4 0.3
#> 5 5.0 3.4 1.5 0.2
#> 6 4.4 2.9 1.4 0.2
left3 <- sqldf("select * from df1 as a left join df2 as b on a.Species=b.Species")
head(left3)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species Sepal.Length
#> 1 5.1 3.5 1.4 0.2 setosa 4.3
#> 2 5.1 3.5 1.4 0.2 setosa 4.4
#> 3 5.1 3.5 1.4 0.2 setosa 4.4
#> 4 5.1 3.5 1.4 0.2 setosa 4.4
#> 5 5.1 3.5 1.4 0.2 setosa 4.5
#> 6 5.1 3.5 1.4 0.2 setosa 4.6
#> Sepal.Width Petal.Length Petal.Width Species
#> 1 3.0 1.1 0.1 setosa
#> 2 2.9 1.4 0.2 setosa
#> 3 3.0 1.3 0.2 setosa
#> 4 3.2 1.3 0.2 setosa
#> 5 2.3 1.3 0.3 setosa
#> 6 3.1 1.5 0.2 setosaNote: 基础包中的merge,当all = F就是内连接,all = T就是全连接,all.x = T就是左联结,all.y = T就是右连接;
Below examples show how to run a variety of data frame manipulations in R without SQL and then again with SQL
subset statment vs where statment
a2r <- subset(CO2, grepl("^Qn", Plant))
head(a2r, 10)
#> Grouped Data: uptake ~ conc | Plant
#> Plant Type Treatment conc uptake
#> 1 Qn1 Quebec nonchilled 95 16.0
#> 2 Qn1 Quebec nonchilled 175 30.4
#> 3 Qn1 Quebec nonchilled 250 34.8
#> 4 Qn1 Quebec nonchilled 350 37.2
#> 5 Qn1 Quebec nonchilled 500 35.3
#> 6 Qn1 Quebec nonchilled 675 39.2
#> 7 Qn1 Quebec nonchilled 1000 39.7
#> 8 Qn2 Quebec nonchilled 95 13.6
#> 9 Qn2 Quebec nonchilled 175 27.3
#> 10 Qn2 Quebec nonchilled 250 37.1
a2s <- sqldf("select * from CO2 where Plant like 'Qn%'")
head(a2s, 10)
#> Plant Type Treatment conc uptake
#> 1 Qn1 Quebec nonchilled 95 16.0
#> 2 Qn1 Quebec nonchilled 175 30.4
#> 3 Qn1 Quebec nonchilled 250 34.8
#> 4 Qn1 Quebec nonchilled 350 37.2
#> 5 Qn1 Quebec nonchilled 500 35.3
#> 6 Qn1 Quebec nonchilled 675 39.2
#> 7 Qn1 Quebec nonchilled 1000 39.7
#> 8 Qn2 Quebec nonchilled 95 13.6
#> 9 Qn2 Quebec nonchilled 175 27.3
#> 10 Qn2 Quebec nonchilled 250 37.1
all.equal(as.data.frame(a2r), a2s)
#> [1] "Component \"Plant\": Attributes: < Component \"levels\": Lengths (3, 12) differ (string compare on first 3) >"
#> [2] "Component \"Type\": Attributes: < Component \"levels\": Lengths (1, 2) differ (string compare on first 1) >"
#> [3] "Component \"Treatment\": Attributes: < Component \"levels\": Lengths (1, 2) differ (string compare on first 1) >"
data(farms, package = "MASS")
a3r <- subset(farms, Manag %in% c("BF", "HF"))
row.names(a3r) <- NULL
a3r
#> Mois Manag Use Manure
#> 1 M1 BF U2 C2
#> 2 M1 HF U1 C2
#> 3 M1 HF U2 C2
#> 4 M1 HF U3 C3
#> 5 M5 HF U3 C3
#> 6 M4 HF U1 C1
#> 7 M2 BF U1 C1
#> 8 M1 BF U3 C1
a3s <- sqldf("select * from farms where Manag in ('BF', 'HF')")
a3s
#> Mois Manag Use Manure
#> 1 M1 BF U2 C2
#> 2 M1 HF U1 C2
#> 3 M1 HF U2 C2
#> 4 M1 HF U3 C3
#> 5 M5 HF U3 C3
#> 6 M4 HF U1 C1
#> 7 M2 BF U1 C1
#> 8 M1 BF U3 C1
identical(a3r, a3s)
#> [1] TRUE
a4r <- subset(warpbreaks, breaks >= 20 & breaks <= 30)
head(a4r, 10)
#> breaks wool tension
#> 1 26 A L
#> 2 30 A L
#> 4 25 A L
#> 8 26 A L
#> 11 21 A M
#> 12 29 A M
#> 17 30 A M
#> 20 21 A H
#> 21 24 A H
#> 25 28 A H
a4s <- sqldf("select * from warpbreaks where breaks between 20 and 30", row.names = TRUE)
head(a4s, 10)
#> breaks wool tension
#> 1 26 A L
#> 2 30 A L
#> 4 25 A L
#> 8 26 A L
#> 11 21 A M
#> 12 29 A M
#> 17 30 A M
#> 20 21 A H
#> 21 24 A H
#> 25 28 A H
identical(a4r, a4s)
#> [1] TRUE
a5r <- subset(farms, Mois == 'M1')
a5r
#> Mois Manag Use Manure
#> 1 M1 SF U2 C4
#> 2 M1 BF U2 C2
#> 5 M1 HF U1 C2
#> 6 M1 HF U2 C2
#> 7 M1 HF U3 C3
#> 11 M1 BF U3 C1
#> 18 M1 NM U1 C0
a5s <- sqldf("select * from farms where Mois = 'M1'", row.names = TRUE)
a5s
#> Mois Manag Use Manure
#> 1 M1 SF U2 C4
#> 2 M1 BF U2 C2
#> 5 M1 HF U1 C2
#> 6 M1 HF U2 C2
#> 7 M1 HF U3 C3
#> 11 M1 BF U3 C1
#> 18 M1 NM U1 C0
identical(a5r, a5s)
#> [1] TRUE
a6r <- subset(farms, Mois == 'M2')
a6r
#> Mois Manag Use Manure
#> 3 M2 SF U2 C4
#> 4 M2 SF U2 C4
#> 10 M2 BF U1 C1
#> 17 M2 NM U1 C0
a6s <- sqldf("select * from farms where Mois = 'M2'", row.names = TRUE)
a6s
#> Mois Manag Use Manure
#> 3 M2 SF U2 C4
#> 4 M2 SF U2 C4
#> 10 M2 BF U1 C1
#> 17 M2 NM U1 C0
identical(a6r, a6s)
#> [1] TRUErbind
a7r <- rbind(a5r, a6r)
a7r
#> Mois Manag Use Manure
#> 1 M1 SF U2 C4
#> 2 M1 BF U2 C2
#> 5 M1 HF U1 C2
#> 6 M1 HF U2 C2
#> 7 M1 HF U3 C3
#> 11 M1 BF U3 C1
#> 18 M1 NM U1 C0
#> 3 M2 SF U2 C4
#> 4 M2 SF U2 C4
#> 10 M2 BF U1 C1
#> 17 M2 NM U1 C0
a7s <- sqldf("select * from a5s union all select * from a6s", row.names = TRUE)
a7s
#> Mois Manag Use Manure
#> 1 M1 SF U2 C4
#> 2 M1 BF U2 C2
#> 5 M1 HF U1 C2
#> 6 M1 HF U2 C2
#> 7 M1 HF U3 C3
#> 11 M1 BF U3 C1
#> 18 M1 NM U1 C0
#> 3 M2 SF U2 C4
#> 4 M2 SF U2 C4
#> 10 M2 BF U1 C1
#> 17 M2 NM U1 C0
identical(a7r, a7s)
#> [1] TRUEaggregate - avg conc and uptake by Plant and Type
a8r <- aggregate(iris[1:2], iris[5], mean)
a8r
#> Species Sepal.Length Sepal.Width
#> 1 setosa 5.006 3.428
#> 2 versicolor 5.936 2.770
#> 3 virginica 6.588 2.974
a8s <- sqldf('
select Species, avg("Sepal.Length") `Sepal.Length`, avg("Sepal.Width") `Sepal.Width`
from iris
group by Species
')
a8s
#> Species Sepal.Length Sepal.Width
#> 1 setosa 5.006 3.428
#> 2 versicolor 5.936 2.770
#> 3 virginica 6.588 2.974
all.equal(a8r, a8s)
#> [1] TRUEby - avg conc and total uptake by Plant and Type
a9r <- do.call(
rbind,
by(iris, iris[5], function(x) {
with(
x,
data.frame(
Species = Species[1],
mean.Sepal.Length = mean(Sepal.Length),
mean.Sepal.Width = mean(Sepal.Width),
mean.Sepal.ratio = mean(Sepal.Length / Sepal.Width)
)
)
})
)
row.names(a9r) <- NULL
a9r
#> Species mean.Sepal.Length mean.Sepal.Width mean.Sepal.ratio
#> 1 setosa 5.006 3.428 1.470188
#> 2 versicolor 5.936 2.770 2.160402
#> 3 virginica 6.588 2.974 2.230453
a9s <- sqldf('
select Species,
avg("Sepal.Length") `mean.Sepal.Length`,
avg("Sepal.Width") `mean.Sepal.Width`,
avg("Sepal.Length"/"Sepal.Width") `mean.Sepal.ratio`
from iris
group by Species
')
a9s
#> Species mean.Sepal.Length mean.Sepal.Width mean.Sepal.ratio
#> 1 setosa 5.006 3.428 1.470188
#> 2 versicolor 5.936 2.770 2.160402
#> 3 virginica 6.588 2.974 2.230453
all.equal(a9r, a9s)
#> [1] TRUEhead - top 3 breaks
a10r <- head(warpbreaks[order(warpbreaks$breaks, decreasing = TRUE), ], 3)
row.names(a10r) <- NULL
a10r
#> breaks wool tension
#> 1 70 A L
#> 2 67 A L
#> 3 54 A L
a10s <- sqldf("select * from warpbreaks order by breaks desc limit 3")
a10s
#> breaks wool tension
#> 1 70 A L
#> 2 67 A L
#> 3 54 A L
identical(a10r, a10s)
#> [1] TRUEhead - bottom 3 breaks
a11r <- head(warpbreaks[order(warpbreaks$breaks), ], 3)
row.names(a11r) <- NULL
a11r
#> breaks wool tension
#> 1 10 A H
#> 2 12 A M
#> 3 13 B H
a11s <- sqldf("select * from warpbreaks order by breaks limit 3")
a11s
#> breaks wool tension
#> 1 10 A H
#> 2 12 A M
#> 3 13 B H
identical(a11r, a11s)
#> [1] TRUEave - rows for which v exceeds its group average where g is group
DF <- data.frame(g = rep(1:2, each = 5), t = rep(1:5, 2), v = 1:10)
DF
#> g t v
#> 1 1 1 1
#> 2 1 2 2
#> 3 1 3 3
#> 4 1 4 4
#> 5 1 5 5
#> 6 2 1 6
#> 7 2 2 7
#> 8 2 3 8
#> 9 2 4 9
#> 10 2 5 10
a12r <- subset(DF, v > ave(v, g, FUN = mean))
row.names(a12r) <- NULL
a12r
#> g t v
#> 1 1 4 4
#> 2 1 5 5
#> 3 2 4 9
#> 4 2 5 10
Gavg <- sqldf("select g, avg(v) as avg_v from DF group by g")
a12s <- sqldf("select DF.g, t, v from DF, Gavg where DF.g = Gavg.g and v > avg_v")
a12s
#> g t v
#> 1 1 4 4
#> 2 1 5 5
#> 3 2 4 9
#> 4 2 5 10
identical(a12r, a12s)
#> [1] TRUEsame but reduce the two select statements to one using a subquery
a13s <- sqldf("
select g, t, v
from DF d1, (select g as g2, avg(v) as avg_v from DF group by g)
where d1.g = g2 and v > avg_v
")
a13s
#> g t v
#> 1 1 4 4
#> 2 1 5 5
#> 3 2 4 9
#> 4 2 5 10
identical(a12r, a13s)
#> [1] TRUEsame but shorten using natural join
a14s <- sqldf("
select g, t, v
from DF
natural join (select g, avg(v) as avg_v from DF group by g)
where v > avg_v
")
a14s
#> g t v
#> 1 1 4 4
#> 2 1 5 5
#> 3 2 4 9
#> 4 2 5 10
identical(a12r, a14s)
#> [1] TRUEtable
a15r <- table(warpbreaks$tension, warpbreaks$wool)
a15r
#>
#> A B
#> L 9 9
#> M 9 9
#> H 9 9
a15s <- sqldf("
select sum(wool = 'A'), sum(wool = 'B')
from warpbreaks
group by tension
")
a15s
#> sum(wool = 'A') sum(wool = 'B')
#> 1 9 9
#> 2 9 9
#> 3 9 9
all.equal(as.data.frame.matrix(a15r), a15s, check.attributes = FALSE)
#> [1] TRUEreshape
DF
#> g t v
#> 1 1 1 1
#> 2 1 2 2
#> 3 1 3 3
#> 4 1 4 4
#> 5 1 5 5
#> 6 2 1 6
#> 7 2 2 7
#> 8 2 3 8
#> 9 2 4 9
#> 10 2 5 10
t.names <- paste("t", unique(as.character(DF$t)), sep = "_")
a16r <- reshape(DF, direction = "wide", timevar = "t", idvar = "g", varying = list(t.names))
a16r
#> g t_1 t_2 t_3 t_4 t_5
#> 1 1 1 2 3 4 5
#> 6 2 6 7 8 9 10
a16s <- sqldf("
select g,
sum((t == 1) * v) t_1,
sum((t == 2) * v) t_2,
sum((t == 3) * v) t_3,
sum((t == 4) * v) t_4,
sum((t == 5) * v) t_5
from DF
group by g
")
a16s
#> g t_1 t_2 t_3 t_4 t_5
#> 1 1 1 2 3 4 5
#> 2 2 6 7 8 9 10
all.equal(a16r, a16s, check.attributes = FALSE)
#> [1] TRUEorder
a17r <- Formaldehyde[order(Formaldehyde$optden, decreasing = TRUE), ]
row.names(a17r) <- NULL
a17r
#> carb optden
#> 1 0.9 0.782
#> 2 0.7 0.626
#> 3 0.6 0.538
#> 4 0.5 0.446
#> 5 0.3 0.269
#> 6 0.1 0.086
a17s <- sqldf("select * from Formaldehyde order by optden desc")
a17s
#> carb optden
#> 1 0.9 0.782
#> 2 0.7 0.626
#> 3 0.6 0.538
#> 4 0.5 0.446
#> 5 0.3 0.269
#> 6 0.1 0.086
identical(a17r, a17s)
#> [1] TRUEcentered moving average of length 7
set.seed(1)
DF <- data.frame(x = rnorm(15, 1:15))
DF
#> x
#> 1 0.3735462
#> 2 2.1836433
#> 3 2.1643714
#> 4 5.5952808
#> 5 5.3295078
#> 6 5.1795316
#> 7 7.4874291
#> 8 8.7383247
#> 9 9.5757814
#> 10 9.6946116
#> 11 12.5117812
#> 12 12.3898432
#> 13 12.3787594
#> 14 11.7853001
#> 15 16.1249309
s18 <- sqldf("
select a.x x, avg(b.x) movavgx
from DF a, DF b
where a.row_names - b.row_names between -3 and 3
group by a.row_names having count(*) = 7
order by a.row_names+0
", row.names = TRUE
)
s18
#> x movavgx
#> 1 5.595281 4.044759
#> 2 5.329508 5.239727
#> 3 5.179532 6.295747
#> 4 7.487429 7.371495
#> 5 8.738325 8.359567
#> 6 9.575781 9.368186
#> 7 9.694612 10.396647
#> 8 12.511781 11.010629
#> 9 12.389843 12.065858
r18 <- data.frame(x = DF[4:12,], movavgx = rowMeans(embed(DF$x, 7)))
row.names(r18) <- NULL
r18
#> x movavgx
#> 1 5.595281 4.044759
#> 2 5.329508 5.239727
#> 3 5.179532 6.295747
#> 4 7.487429 7.371495
#> 5 8.738325 8.359567
#> 6 9.575781 9.368186
#> 7 9.694612 10.396647
#> 8 12.511781 11.010629
#> 9 12.389843 12.065858
all.equal(r18, s18)
#> [1] TRUEmerge. a19r and a19s are same except row order and row names
A <- data.frame(a1 = c(1, 2, 1), a2 = c(2, 3, 3), a3 = c(3, 1, 2))
B <- data.frame(b1 = 1:2, b2 = 2:1)
a19s <- sqldf("select * from A, B")
a19s
#> a1 a2 a3 b1 b2
#> 1 1 2 3 1 2
#> 2 1 2 3 2 1
#> 3 2 3 1 1 2
#> 4 2 3 1 2 1
#> 5 1 3 2 1 2
#> 6 1 3 2 2 1
a19r <- merge(A, B)
a19r
#> a1 a2 a3 b1 b2
#> 1 1 2 3 1 2
#> 2 2 3 1 1 2
#> 3 1 3 2 1 2
#> 4 1 2 3 2 1
#> 5 2 3 1 2 1
#> 6 1 3 2 2 1
Sort <- function(DF) {DF[do.call(order, DF),]}
all.equal(Sort(a19s), Sort(a19r), check.attributes = FALSE)
#> [1] TRUER interact with Linux command
system('mkdir ./data/folder1')
system('cd ./data/;cp read_sql.csv ./folder1')
system("cd ./data/;echo 'echo test'> echo.sh")
system('cd ./data/;mv echo.sh echo2.sh')
system('rm -rf ./data/folder1')